Surveillance of Retroelement Expression and Nucleic-Acid Immunity by Histone Methyltransferase SETDB1
Abstract
In human cancers, histone methyltransferase SETDB1 (SET domain, bifurcated 1) is frequently overexpressed but its significance in carcinogenesis remains elusive. A recent study shows that SETDB1 downregulation induces de-repression of retroelements and innate immunity in cancer cells. The possibility of SETDB1 functioning as a surveillant of retroelement expression is discussed in this study: the cytoplasmic presence of retroelement-derived nucleic acids (RdNAs) drives SETDB1 into the nucleus by the RNA-interference route, rendering the corresponding retroelement transcriptionally inert. These RdNAs could, therefore, be signals of genome instability sent out for SETDB1 present in the cytoplasm to maintain genome integrity.
1 Introduction
SETDB1 catalyzes the synthesis of trimethylated histone H3 lysine 9 (H3K9me3), a repressive mark to which heterochromatin protein 1 (HP1) is recruited and deposited, thus inducing the formation of heterochromatin in the region. SETDB1 also participates in transcriptional repression of target gene promoters. Many transcription factors, including ERG,1 mAM,2 KAP1,3 DNMT3A,4 SP3,5 OCT4,6, 7 MBD1/CAF1,8 MTA2,9 and SIN3A9 have target genes under the regulatory control of SETDB1. However, SETDB1 does not function exclusively in the nucleus; it also exists dynamically in the cytoplasm10, 11 depending on the cell type and level of expression.11 In reality, SETDB1 has both nuclear localization (NLS) and a nuclear export signal (NES), signifying the role of SETDB1 as a nucleocytoplasmic shuttle protein.12 Even though the cellular processes in which SETDB1 shuttling takes part in are yet to be elucidated, this suggests that SETDB1 be considered more than a simple, nuclear H3K9me3 writer. With regard to protein structure, SETDB1 has several features that other histone methyltransferases do not have, potentially endowing SETDB1 with certain singular functions. SETDB1, along with SETDB2, are the only histone methyltransferases with a bifurcated catalytic SET domain, which has been conserved from flies to worms to humans.12 The contribution of the SET domain-splitting (i.e., bifurcating) 347-amino acid spacer (343 aa in mice) to the SETDB1 functions remains unknown. In addition to the SET domain, SETDB1 has two consecutive Tudor domains and a putative methyl-binding domain (MBD) in the amino terminal, likely functioning in methyl-lysine13 and methyl-cytosine binding, respectively, but their bona fide functions are yet to be established. For instance, which lysine residues (on histones or non-histone proteins) and which state (mono-, di-, or tri-) of lysine methylation the SETDB1-Tudor can specifically bind is unknown, and unlike the MBDs of MeCP2 and MBD1, SETDB1-MBD has not conclusively been described to selectively bind methylated CpG regions. Exploring the individual roles of these domains is necessary to expand our comprehension of the pleiotropic functions of SETDB1.
SETDB1 is known to play important roles in early development and embryonic stem cell self-renewal including the cell-lineage (trophectoderm vs. inner cell mass) specification at the blastocyst stage6, 7, 14 and the blastocyst-derived embryonic stem cell population15 which has been recently reviewed12 and consequently, not discussed in this review. SETDB1 is also known as an indispensable structural constituent of promyelocytic leukemia-nuclear body (PML-NB), a large proteinaceous structure through which many proteins, involved in a variety of biological processes, transiently associate.16 In addition to its structural contribution to PML-NBs, SETDB1 also controls transcriptional repression of genes associated with PML-NBs.17 Additionally, discussed at length below, SETDB1 suppresses endogenous retroviruses (ERVs) when associated with KRAB-ZNF/KAP1 complexes18 in embryonic stem cells,19 which, as recently suggested,20 could explain the frequent upregulation of SETDB1 in cancer cells. Seemingly involved in various cellular processes, the role SETDB1 plays in these processes has only separately been discussed, with little assimilation. A variety of studies over the 16 years since the discovery of SETDB1 as a histone methyltransferase and transcriptional repressor1 has accumulated a massive wealth of information. This review aims to integrate these significant findings into a single big picture overview, containing the features of SETDB1 expression in cancer cells and its oncogenic role in tumorigenesis, the molecular functions, with a focus on mechanistic aspects, of SETDB1 in cancers, and finally, a discussion on the possibility of SETDB1 as a surveillant of retroelement expressions acting through RNA interference.
2 SETDB1 as a Bona-Fide Oncoprotein
An increasing volume of recent evidence suggests that SETDB1 plays an important role in tumorigenesis of various cancers, validating the classification of SETDB1 as an oncoprotein. The conspicuous presence of SETDB1 is observable from gene copy amplification in tumors where SETDB1 is overexpressed. This relationship is outlined in several case studies reporting the involvement of SETDB1 in cancer recurrence, suggesting SETDB1 overexpression as a promising prognostic marker and target in future chemotherapeutic strategy.
2.1 SETDB1 Gene Amplification in Cancer Cells
The SETDB1 gene is located in the 1q21 interval of the human chromosome, a region known to be recurrently amplified in a variety of solid tumors.21 The SETDB1 gene, consequently, is observed to have multiple copies in various cancers including melanomas22 non-small and small cell lung cancers (NSCLC and SCLC23), hepatocellular carcinomas (HCC),24 and breast cancers.25 In HCC, for example, a moderate copy gain of the SETDB1 gene was found in about 70% or more samples, and this gain intimately associated with a corresponding increase in SETDB1 expression.24 From meta-analysis of 958 public breast-cancer genome sequencing data, SETDB1 gene amplification was found to have the highest frequency in basal-like subtype (>30%)26 the most aggressive breast cancer subtype associated with higher rates of metastasis and death.27 Analysis of public cancer data (cBioPortal; http://www.cbioportal.org), reveals a high frequency (>8%) of SETDB1 gene amplification in breast, liver, lung, and esophagus-stomach cancers compared to colorectal, kidney, and prostate cancers (Figure 1A). In comparison, EZH2, the H3K27me3 writer, has been previously established as an oncogene in breast and prostate cancers28 with elevated levels of EZH2 detected in more advanced stages of cancers.29 However, EZH2 has not shown as much copy gain in cancers as SETDB1 has (Figure 1A). Meanwhile, SETDB1 loss-of-function mutations have been frequently observed in mesothelioma although the causal relationship between SETDB1 deletion and oncogenesis is unknown.30
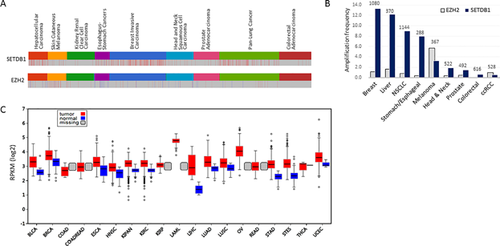
2.2 SETDB1 Overexpression in Cancer Cells
When the genes present in the recurrently amplified 1q21 interval of the chromosome were individually and forcibly expressed in zebrafish, one of the genes within the region, SETDB1, could significantly accelerate the onset of melanoma.22 This is the first direct demonstration of the strong correlation between SETDB1 overexpression and cancer development and the role of SETDB1 as an oncogene. Analysis of public transcriptome data proves that SETDB1 overexpression is common in a variety of cancers and does not necessarily require gene amplification (Figure 1B). For example, SETDB1 copy gain is a rare event in colorectal and kidney cancers, but SETDB1 expression is still much higher when compared to normal samples. The mechanisms involved in upregulating SETDB1 in these cancers have yet to be identified. A noteworthy finding from these data are the observed overexpression of SP1, which transcriptionally regulates SETDB1 by binding at the promoter region, in HCCs, which is a plausible mechanism of SETDB1 upregulation.24
From SETDB1 studies in different cancers, it is ostensive how certain oncophenotypes are found to be linked with SETDB1 overexpression. Increased expression of SETDB1 was studied in NSCLC samples where it was observed that forced SETDB1 expression raised their clonogenicity and tumor size in a xenograft model, that is, it appeared to be positively correlated with severity of the disease phenotype.23, 31 In another study of NSCLC, 19% of stage-I tumor samples were found to overexpress SETDB1, but in contrast to the ERCC1, RRM1, and BRCA1 markers for advanced-stage NSCLC patients,32 SETDB1 overexpression was proposed as a significant prognostic marker to predict tumor recurrence in early stage NSCLC patients.33 In hepatocellular carcinoma (HCC) where the SETDB1 gene is frequently amplified, SETDB1 was significantly upregulated, just as in all its metastatic foci in different organs,24 strongly hinting at the significance of SETDB1 overexpression in HCC metastatic progression. Another study with public HCC data revealed that later-stage cancers express higher levels of SETDB1,34 showing a strong correlation between SETDB1 expression level and the grade of tumorigenesis. SETDB1 overexpression was also found in other solid cancers including glioma,35 breast,26 and prostate cancers.36
2.3 Inhibition of Tumor Growth by SETDB1 Repression
In most cases, SETDB1 knockdown reduces the tumorigenic and metastatic potential of cancer cells by diminishing the ability of proliferation, migration, and invasion, which are demonstrated in a variety of cancers including prostate cancer,35 HCC,24 NSCLC,31 breast cancer,25 and glioma cells.36 One such instance of endogenous miRNA-mediated interference of SETDB1 expression is in breast cancer, where microRNA-7 (mir-7) can inhibit metastasis of cancer cells and decrease breast cancer stem cells by targeting 3′-UTR of the SETDB1 transcript.37 Mir-29a, with the potential to target SETDB1 mRNA, has been suggested as a negative regulator for SETDB1,24 and is frequently downregulated in human HCC.38 Mithramycin, which suppresses the promoter activity of SETDB1 by inhibiting the binding of SP1 transcription factor,39 hinders the growth of SETDB1-amplified cells compared to cells with normal SETDB1 level.23 However, careful interpretation of results is necessary, given the multiplicity of SP1 target genes and the possibility of off-target effects from mithramycin treatment. Collectively, these observations prove that SETDB1 is a bona fide oncogene, essential for cancer cell proliferation and metastasis, and suggest the potential of SETDB1 as a promising target for new chemotherapeutic strategies.
3 Molecular Functions of Oncogenic SETDB1 in Cancer Cells
SETDB1 studies in a variety of cancers have consistently pointed to its oncogenic role. However, the majority of these findings have been derived from phenotypic observations in SETDB1 overexpression or knockdown experiments. Owing to the wide range of epi-driver protein targets and their global effects, these findings are mostly indirect and circumstantial, while the detailed molecular mechanism of SETDB1-involved oncogenesis remains largely unknown. However, considering that a number of different cancers respond in a similar manner to the activity of SETDB1, SETDB1 may share a common oncogenic role in these cancers.
3.1 Repression of Tumor Suppressors?
Generally, SETDB1, combined with chromatin remodelers, render target gene promoters transcriptionally inert, thereby regulating cellular gene expression in response to environmental stimuli. An easily conceivable scenario for the oncogenic mechanism of SETDB1 can be that SETDB1 may exercise its influence through the silencing of tumor suppressor p53 at the transcriptional level. This hypothesis, however, has gained no experimental support at present. Standing opposite to the hypothesis, Noh et al. observed that p53 could suppress SETDB1 expression. This study showed that paclitaxel (PTX), which induces both p53 and p21,40 repressed SETDB1 expression in lung cancer cells, and p53 directly inhibited SETDB1 transcription by binding to the SETDB1 promoter.41 In another study, Fei et al. concluded that SETDB1 could increase the stability of p53 R249S, an already stable gain-of-function mutation,42 by dimethylating K370, also enhancing its oncogenic activity.34 The same paper observed that SETDB1 also methylated wild-type p53, which however, had little effect on cancer cell growth because of its intrinsic instability. In the study using zebrafish melanoma model (see section 2.2), the oncophenotype caused by SETDB1 overexpression was shown to be largely different from the p53 knockout phenotype,22 which refutes the possibility of a cause-and-effect interaction between SETDB1 and p53. In summary, it has not been evidenced that SETDB1 transcriptionally regulates p53 in cancer cells, and the observations from the above-described studies offer more disapproval than they do support the scenario.
3.2 Evading Apoptosis by SETDB1 Amplification
In a different scenario, SETDB1 may contribute to tumor growth by repressing a group of genes implicated in the apoptosis of cancer cells. SETDB1 is involved in spermatogenesis in mice. It has been shown that SETDB1 knockdown induces apoptosis of spermatogonial stem cells,43 which was explained in a later study by a consequential increase in expression of downstream pro-apoptotic genes, Bim and Puma.44 It was later shown that SETDB1, collaborating with AKT,45 inhibits the transcriptional regulatory function of FOXO1,44 and contributes to cell survival by repressing multiple apoptotic genes.46 This is reminiscent of the apoptotic phenotype after SETDB1 knockdown in a variety of cancer cells including glioma cell lines.35 These studies suggest a curious link between SETDB1 interaction and the PTEN/AKT/FOXO1 signaling pathway in cancer cells. Given this anti-apoptotic function of SETDB1 under typical conditions, SETDB1 upregulation may allow greater ease for cancer cells to circumvent apoptotic conditions, effectively evading the threat of programmed cell death. Another possibility considers the ability of SETDB1 to bind and methylate p53 post-translationally,34 which upon overexpression of SETDB1 could excessively methylate otherwise non-target lysine residue(s) on p53, hence destabilizing p53 proteins and inhibiting apoptosis in cancer cells. However, such a case has not been reported thus far.
3.3 Stabilizing Cancer Stem Cells
SETDB1 overexpression may contribute to oncogenesis by stabilizing cancer stem cells. There are several cases of supporting evidence for this theory. Setdb1 deletion causes a rapid depletion of hematopoietic stem and progenitor cells (HSPCs), indicating that SETDB1 is essential for the maintenance of HSPSs in mice.47 MiR-7, by direct inhibition of SETDB1, reverses the epithelial-mesenchymal transition (EMT) of breast cancer stem cells.37 SETDB1 interacts with ΔNp6325 which is implicated in the self-renewal of breast cancer stem cells and control of mammary cancer stem cell homeostasis.48 Stemness-related functions of SETDB1 are better known in embryonic stem cells (ESCs),12 where Setdb1-null blastocysts fail to produce ESCs de novo.15 SETDB1 interacts with OCT4, and SETDB1 knockdown in ESCs markedly alters the colony morphology and increases expression of differentiation marker genes, which are normally repressed by OCT4-guided SETDB1.6, 7, 14 In fly, SETDB1 regulates the formation of pericentric heterochromatin in germarial germline stem cells, which generates oocytes.49 It has also been established that SETDB1 is necessary for mammalian germline development.50 Given its functions in embryonic, germline, and cancer stem cells, SETDB1 could be considered as having a general role in maintaining homeostasis and self-renewal in cells with features of stemness.
4 SETDB1 as Retroelement Silencer
The important function of SETDB1 in repression of retroelements has been well established. The transcriptional repression is accomplished either by SETDB1 alone or in combination with DNA methyltransferases (DNMTs) depending on the type of cell. In this section, the dynamics of RdNAs, the related processes that ultimately elicit innate immunity in cancer cells, and the involvement of SETDB1 in retroelement biology are discussed.
4.1 Types of Endogenous Retroelements and Their Replication
Endogenous retroelements constitute up to 43% of the human genome and are classified into two broad categories: the LTR-bound retroelements characterized by the presence of long-terminal repeats (LTRs), and the non-LTR retroelements that lack LTR. There are millions of LTR and non-LTR retroelement copies in the genome. However, complete copies with all ORFs predicted to be intact are approximately 100 or less in number,51 and not necessarily active or infectious as these capabilities depend on the type and level of epigenetic silencing in the cells. Long and short interspersed nuclear elements (LINEs and SINEs, respectively) are non-LTR retroelements. Intact LINE1 retroelements produce two functional proteins, ORF1p, an RNA-binding protein and ORF2p, having both endonuclease and reverse transcriptase (RTase) activity. These proteins bind LINE1 RNA and enter the nucleus, facilitating insertion of a new LINE1 copy into the genome.52 Although the vast majority of LINE1 sequences in the human genome are retrotransposition-defective, retrotransposition is frequently observed in pathophysiological conditions such as cancer.53
LTR elements comprise endogenous retroviruses (ERVs) and mammalian apparent LTR retrotransposons (MaLRs).54 The basic structure of an ERV genome consists of Gag, Pol, Pro, and Env ORFs in its simplest form. Of these ERV genes, the Pol gene encodes a pol polyprotein containing the protease (PR), RTase/RNase H, and integrase (INT), which are the molecular tools enabling retrotransposition. ERV virions contain the genome, consisting of two copies of viral single-stranded RNA (ssRNA) which are reverse-transcribed by the encapsidated RTase in the cytoplasm of an infected cell after uncoating.55 The resulting cDNA, still in complex with viral proteins, enters the nucleus to integrate into host DNA. During the viral life cycle, various RdNA species can be produced in different intracellular compartments.
4.2 Retroelement Biology and Innate Immunity
Endogenous retroelements possess retroviral features such as the ability to reverse-transcribe their RNA genome into DNA and to form virions, which do not go unnoticed by the immune system. RdNAs too, both final and intermediate, that are produced during replication of endogenous retroelements could be considered immunogenic.56 LINEs, for example, can produce double-stranded RNAs (dsRNAs) from their bidirectional promoters at 5′-UTR,57 and ERVs can churn out endogenous retroviral DNA into the cytoplasm during reverse transcription. Failure to remove these atypical nucleic acids by metabolism causes an inappropriate activation of nucleic acid sensing pathways and a resulting type I interferon (IFN) response.58 Certain specialized innate receptors or sensors that defend against exogenous retroviruses can also detect the immunogenic RdNAs.56 The innate nucleic acid receptors, each with unique substrate specificity, include the dsRNA sensor toll-like receptor 3 (TLR3), the ssRNA sensor TLR7, the unmethylated CpG DNA sensor TLR9, the cytosolic RNA helicases RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated gene 5), and the dsDNA sensor cyclic GMP-AMP synthase (cGAS), all of which elicit type I IFN signaling via direct activation of STING (stimulator of IFN genes).59
Mechanistically, two nucleic acid-sensing pathways mediate the detection of RNA or DNA by the innate immune system. RNA sensing is carried out by RIG-I and MDA5, which detect unique structures of viral RNA.60 Upon stimulation, these sensors bind to MAVS (mitochondrial antiviral signaling) protein on the surface of mitochondria, which in turn relays the signal to TBK1 (TANK-binding kinase 1) and IRF3 (IFN regulatory factor 3), resulting in the production of type I IFNs. During DNA sensing, the cytosolic AIM2 (absent in melanoma 2) receptor detects long dsDNA and stimulates the ASC inflammasome.61 Another dsDNA sensor cGAS, which catalyzes the production of the second messenger cGAMP, activates STING on endoplasmic reticulum, which cascades to the TBK1-IRF3 pathway for induction of an antiviral response.62
4.3 Silencing of Retroelements by SETDB1
Normally, endogenous retroelements are transcriptionally repressed by powerful epigenetic mechanisms, with a different silencing mechanism for each retroelement. Early studies on the expression of intracisternal A particle (IAP) sequences, a type-II ERV in mice, identified DNA methylation as the major suppression mechanism for them. IAPs in mice were derepressed in the DNMT1-deficient fetal somatic cells,63 DNMT3L-deficient perinatal testes,64 and DNMT3A-deficient germ cells.65 IAP was not reactivated in DNMT1-deficient E13.5 primordial germ cells (PGCs),63 which indicate an alternative silencing mechanism in these specialized cells.
It was later identified that LINE1 and type-II ERVs, including IAPs and ETn elements in E13.5 PGCs, are repressed by deposition of SETDB1-catalyzed H3K9me3.50 These observations, therefore, suggest that SETDB1 is an essential guardian against proviral expression prior to the re-establishment of DNA methylation in the germline,50 after DNA methylation reprogramming.66 In addition, in embryonic stem cells but not embryonic fibroblasts, SETDB1 marks and silences class I and class II ERVs.19 This silencing is established independently of DNA methylation in embryonic stem cells.67 The silencing mechanism appears to be cell-type specific because SETDB1 depletion de-represses ERVs in B cells,68 while not in HSPCs in mice.47 In general, the 5′-UTR near the LTR in the IAP element is recognized69 by KRAB-ZNF which, along with KAP1, recruits SETDB1 to mark H3K9me3.3 Transcription co-factor heterogeneous nuclear ribonucleoprotein K (hnRNP K) is recruited to ERVs, depending on KAP1, while SETDB1 binding to these elements requires hnRNP K.70 Class III ERVs such as MERVL elements are silenced by a repressive complex comprising KAP1 and hnRNP K, but differing from class I and II ERVs, class III ERVs are marked by G9a/GLP-deposited H3K9me2.71 Besides ERVs, SETDB1 transcriptionally represses LINE-1 expression in mouse PGCs,50 and protects surviving human cancer cells from lethal drug exposure.72
5 Tripartite Talks in Cancer Cells – Retroelements, SETDB1, and Immunity
Genomic instability, a major driving force in tumorigenesis,73 can be induced by dysregulation of epigenetic mechanisms. The acquisition of genetic and epigenetic mutations can in turn probably induce the silencing mechanism for TSGs and/or the activation mechanism for oncogenes in a random fashion.74 Altered expression levels of global chromatin modifiers such as SETDB1 and EZH2 can be responsible for the generation of tumor-progenitor cells or tumor-initiating cells with stemness (TICs; Figure 2).75 EZH2 overexpression results in impairment of DNA damage repair and expansion of breast TICs,76 and many examples of SETDB1 knockdown and overexpression relating to development, progression, and recurrence of a variety of cancers have already been discussed above. Of note is a worthwhile report on the requirements of SETDB1 in cells like TICs that are experiencing biochemical and physiological transitions. During somatic cell reprogramming, the chromosomal H3K9me3 deposited by SETDB1 serves as an “epigenetic barrier,” and the efficiency of reprogramming is increased by SETDB1 repression.77 Both these observations, namely, the requirement of lower SETDB1 activity in reaching stemness during reprogramming, and higher SETDB1 levels in stably maintaining self-renewal in cancer stem cells (see section 3.3), appear incompatible prima facie. However, if we consider SETDB1 as the “keeper” of a present cellular state or the “resister” for change, they no longer seem incongruous. Bearing the pose of maintaining a steady-state level of SETDB1, a somatic cell can endure the pressure of reprogramming, and a cancer stem cell can easily retain its present state of stemness. In this instance, downregulation of SETDB1 can, for the somatic cell, pose a short-cut route toward stemness (and also toward TICs), while the same downregulation could, to an existing cancer stem cell, present a way of losing stemness toward differentiation. It is known that a similar stemness-acquiring process of cellular reprogramming occurs during the process of transformation into TIC.78
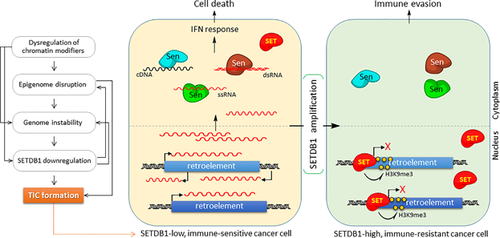
5.1 Scenario for Selective SETDB1 Amplification in Proliferating Cancers
In certain TICs where SETDB1 is expressed at low levels, endogenous retroelements may be derepressed (Figure 2). They are otherwise removed by AGS (Aicardi-Goutieres syndrome) enzymes such as TREX1, SAMHD, RNase H, and cGAS.79 When the amount of cytoplasmic RdNAs has accumulated to a threshold, perhaps through transcriptional activation, inactivation of the AGS enzymes, or a hyperactive RTase, they can be recognized by nucleic acid sensors patrolling the cytoplasm. This, in turn, induces type I IFN response by upregulation of IFN response genes, with the host cell then becoming the target of an immune reaction.20 In cancer cells, this nucleic-acid immunity can be a chronic stress that holds back expansion of the cancer cell population, the cellular response to which may constitute augmenting SETDB1 expression epigenetically or more directly, an increase in SETDB1 copy number. Increased SETDB1 activity, in turn, rehabilitates heterochromatin at retroelements to repress their expression tightly, which enables host cells to sidestep the innate immune response. The remaining few cells that have overcome the stress in these ways are fit to survive and expand infinitely. Based on this scenario, comprising SETDB1, retroelements, and innate immunity, increased SETDB1 expression precedes the whole timeline of cancer development, recurrence, and/or metastasis. In agreement, recent studies have considered SETDB1 as a relevant marker to predict tumor recurrence33, 72 and metastasis.24
5.2 Hypothetical Mechanism of Retroelement Silencing by SETDB1 Via RNA Interference Pathway
SETDB1 is an “amphibious” protein, existing both in the cytoplasm and the nucleus.10, 11, 14 This is evident when SETDB1 is overexpressed in cultured cells; the majority of exogenous SETDB1 protein is pumped out into the cytoplasm. The exclusion of SETDB1 from the nucleus can be important for maintaining a homeostatic balance among different but associated chromatin-modifying proteins, as well as for preserving the epigenomic integrity from excess SETDB1 activity in the nucleus. Once exported, SETDB1, rather than being idle, is expected to function in the cytoplasm, although its specialized functions remain unknown. One fascinating hypothesis is that SETDB1 is involved in the surveillance of the RdNAs in the cytoplasm, an outcome of epigenetic dysregulation of the genome (Figure 3). SETDB1 may have a direct affinity for, or indirectly associate via an intermediator with, those RdNAs. In contrast to typical nucleic-acid sensors, which induce innate and adaptive immune reactions against host cells, SETDB1 may, after associating with the RdNAs, translocate into the nucleus and transcriptionally repress corresponding retroelements (however, the RNA-dependency of SETDB1 trafficking into the nucleus has yet to find support). In this case, the delivered RdNA may serve as a guide, targeting SETDB1 to retroelement loci, transcribing the complement nascent RNA. This scenario reminisces of transcriptional gene silencing (TGS) in RNA interference (RNAi).80, 81
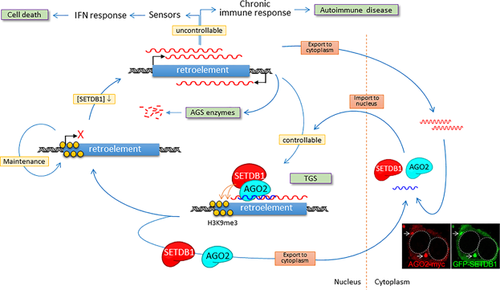
In reality, RNAi pathways are known to be involved in combating endogenous retrotransposon activities in human cells,57 and thus, considered a type of nucleic acid immunity mechanism.59 It is, therefore, interesting to notice that SETDB1 plays an essential role in small RNA-induced TGS via ARGONAUTE 2 (AGO2) association.9 In contrast to post-transcriptional gene silencing (PTGS) in which AGO2 protein forms the RNA-induced silencing complex (RISC) and targets mRNAs sequences for degradation in the cytoplasm, RNAi-mediated TGS can trigger chromatin modifications that lead to heterochromatinization of target loci in the nucleus.81, 82 Both SETDB1 and AGO2 were observed to interact with non-coding RNA transcribed at the target promoter, recruit the SIN3A-HDAC complex and EZH2, and adorn the promoter with H3K9me3 and H3K27me3 marks to make the region transcriptionally inert.9 Similarly, the RdNA-guided SETDB1-AGO2 complex can scan whole chromosomes in search of loci where retroelements are actively transcribed, and finally install a repression mechanism. As previously observed, the cytoplasmic localization of SETDB1 is cell-type dependent.11 In addition, there is a possibility that, differing from the RdNAs, exogenous siRNAs can elicit a distinct TGS mechanism. Considering these, the observation of the nucleus-restricted SETDB1-AGO2 interaction during TGS in T47D cells9 can be reconciled with the present hypothesis. An interesting possibility is that SETDB1 can, through localizing at PML-NB6, 17 where higher-order chromatin loops are hooked83 and a crowd of retroelements collectively present themselves to nucleoplasm, simultaneously access and leaf through a large fraction of the genome quickly. Notably, the increase in the number and size of PML-NBs was seen after viral infection and IFN treatment.84 If this is the case, the ability to utilize PML-NB can be additional benefit of SETDB1 involvement in the feedback regulation of retroelements.
Remarkably, retroelement-mediated innate immunity can be a serious issue in some autoimmune diseases. The same RdNA sensors that shield from viral infection can cause autoimmune diseases as exemplified by the involvement of TLRs in the pathologic response to self-DNA/RNAs.79, 85 AGS is another autoimmune disease caused by mutation in enzymes such as TREX1,86 SAMHD,87 and RNase H88 that prevent immunogenic RdNAs from accumulating in the cytoplasm. While cancer cells frequently make use of SETDB1 overexpression to evade innate immunity, cases of autoimmunity behold the accumulation of self RdNAs possibly from SETDB1 downregulation, the possibility of which has yet to be tested. Will the AGS autoimmune symptoms be relieved if SETDB1 is forcibly overexpressed? It may not be that simple as long as SETDB1 assumes a double-edged sword. The therapeutic use of SETDB1 expression in autoimmune diseases prevents the accumulation of self RdNAs, but may also be lending a hand to TICs being released from restraints set by innate immunity. This disconcerting situation highlights the importance of regulating appropriate SETDB1 activity essential for therapeutic intervention aimed at SETDB1. When left to a cell's own unaffected devices to maintain intracellular balance of SETDB1 activity, instead of claiming that SETDB1 controls retroelements, it may be more precise to mention that retroelements regulate the SETDB1 level in the nucleus by adjusting the amount of cytoplasmic RdNAs they produce; the increased cytoplasmic RdNA levels, in turn, lead to uptake of SETDB1 to replenish its reservoir in the nucleus.
6 Conclusions and Outlook
The control of immunity through regulation of retroelement expression in cancer is an emerging research field, and SETDB1 has been discovered as the first key regulator to suppress innate immunity by limiting the overall abundance of retroelement expression in cancer cells.72 The hypothetical scenario of negative feedback regulation of retroelements by SETDB1 exploiting the RNAi-TGS mechanism is fascinating and can be an example of an inter-compartmental, trans-functional RNAi phenomenon. However, as it currently stands, with many unknowns and a dearth of evidence, a lot of experimental work needs to be carried out to verify likely scenarios and test hypothesized mechanisms. The questions that arise in view of the hypothesis are as follows: what is the mechanism by which the SETDB1 is imported into the nucleus? Will the SETDB1-AGO2 interaction be conserved in normal and cancer cells? What will be the fate of SETDB1-AGO2 complex after completion of TGS? Is there a mechanism of pumping out residual SETDB1 proteins to the cytoplasm to maintain a balance between SETDB1 activity in the nucleus and cytoplasm? Finally, it has been well known that epigenetic mutation (epimutation) as well as genetic mutation are the primary causes of initiation, progression, and recurrence of cancers. SETDB1 is attracting increasing attention for brain disorders including schizophrenia,89 autism,90 and Huntington's disease39 as well as cancers and autoimmune diseases. Unlike genetic mutations, mutations of epigenetic origin can, fortunately, be reversed to a normal state. If the questions on how SETDB1 acts in diseased cells and what the target(s) of SETDB1, can be answered, it will undoubtedly be possible to design a therapeutic plan for multiple targets, and at multiple levels in the SETDB1 pathogenic pathway.
Abbreviations
AGS, Aicardi-Goutieres syndrome; ERV, endogenous retrovirus; ESC, embryonic stem cell; HCC, hepatocellular carcinomas; IAP, intracisternal A particle; LINE and SINE, Long and short interspersed nuclear element; LTR, long-terminal repeat; MBD, methyl-binding domain; NLS/NES, nuclear localization/export signal; NSCLC and SCLC, non-small and small cell lung cancers; PGC, primordial germ cell; PML-NB, promyelocytic leukemia-nuclear body; PTGS, post-transcriptionalgene silencing; RdNA, retroelement-derived nucleic acids; RISC, RNA-induced silencing complex; TGS, transcriptional gene silencing; TIC, tumor-initiating cells; TSG, tumor suppressor gene.
Acknowledgements
The author thanks S. Jeong for critical reading of the manuscript and B. Min for obtaining bioinformatics data. This work was supported by grants from the National Research Foundation of Korea (2011-0030049), KIOM (K16130) and KRIBB Initiative Program.
Conflict of Interest
The authors declare no conflict of interest.




