Real-Time pH-Dependent Self-Assembly of Ionisable Lipids from COVID-19 Vaccines and In Situ Nucleic Acid Complexation
Graphical Abstract
The millisecond kinetic evolution of the ordered structure formation upon the complexation of ionisable lipid (SM-102), a novel structural-forming helper lipid (monoolein, MO) and nucleic acids (e.g., mRNA) are quantitatively investigated using in situ time-resolved small angle X-ray scattering coupled with rapid flow mixing, which will help deciphering the optimal ionisable lipid nanoparticle nanostructures for efficient gene delivery.
Abstract
Ionisable amino-lipid is a key component in lipid nanoparticles (LNPs), which plays a crucial role in the encapsulation of RNA molecules, allowing efficient cellular uptake and then releasing RNA from acidic endosomes. Herein, we present direct evidence for the remarkable structural transitions, with decreasing membrane curvature, including from inverse micellar, to inverse hexagonal, to two distinct inverse bicontinuous cubic, and finally to a lamellar phase for the two mainstream COVID-19 vaccine ionisable ALC-0315 and SM-102 lipids, occurring upon gradual acidification as encountered in endosomes. The millisecond kinetic growth of the inverse cubic and hexagonal structures and the evolution of the ordered structural formation upon ionisable lipid-RNA/DNA complexation are quantitatively revealed by in situ synchrotron radiation time-resolved small angle X-ray scattering coupled with rapid flow mixing. We found that the final self-assembled structural identity, and the formation kinetics, were controlled by the ionisable lipid molecular structure, acidic bulk environment, lipid compositions, and nucleic acid molecular structure/size. The implicated link between the inverse membrane curvature of LNP and LNP endosomal escape helps future optimisation of ionisable lipids and LNP engineering for RNA and gene delivery.
Introduction
The unprecedented fast approval of the COVID-19 mRNA vaccines to control the world pandemic has prompted great interest and effort to develop lipid nanoparticle (LNP) systems as nanocarriers for gene therapy.1-3 In the mRNA-LNP COVID-19 vaccines, the main components consist of helper lipids (phospholipids and cholesterol), PEGylated lipids as particle stabilisers, and most importantly ionisable amino-lipids,3-5 which are the key functional components for stabilising and loading the mRNAs. Another key function of these ionisable lipids is to facilitate cellular uptake, endosomal destabilisation, and cytoplasmic release of the mRNAs.6, 7 Remarkably, the two mainstream mRNA COVID-19 vaccines developed by different pharmaceutical companies use structurally similar ionisable lipids, ALC-0315 for the BioNTech/Pfizer vaccine and SM-102 for the Moderna vaccine (Figure 1a), both possessing a multi-tail structure of several saturated aliphatic chains linked to a tertiary amine group via ester bonds. More ionisable lipids sharing similar molecular structural features are also used, e.g. DLin-MC3-DMA in the first FDA-approved siRNA drug Onpattro®, and in on-going investigations.8-10
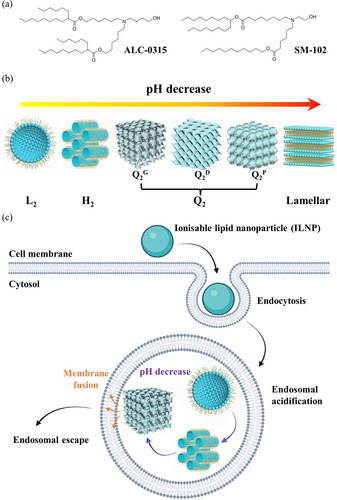
(a) Molecular structures of ionisable lipids, ALC-0315 and SM-102. (b) Schematic diagram of the mesophase transition of ILNPs as the pH decreasing, from L2, H2, Q2, to lamellar. (c) A mechanistic scheme of endosomal escape ability of ILNPs via phase transition and membrane fusion mechanism during endosomal acidification process.
Despite the huge clinical success, the understanding of the dynamic self-assembly structural transformations of the ionisable lipids upon complexation with RNA therapeutics during microfluidic-based LNP synthesis remains largely unreported. Regarding intracellular mechanisms, the exact nature of the interactions between mRNA-LNPs and cell/endosomal membranes has not yet been elucidated. The lack of knowledge on the structural evolution relevant to the LNP formulation process, stability, and functionality at biological interfaces impedes future optimisation of RNA therapeutics.
In 1997, Radler et al. was among the first to report that DNA complexation with cationic liposomes led to a structural transition from liposomes to optically birefringent liquid-crystalline multi-layer structures detected by synchrotron small angle X-ray scattering experiments (SAXS).11 Later the same research group reported an inverse hexagonal (H2) phase of the DNA-liposome complex with improved transfection efficiency over the lamellar structure of the complex.12 Since then, rising evidence showing the existence of non-lamellar inverse structures of LNP delivery vehicles has become available.13-16 Recently, Arteta et al. used SAXS to demonstrate that a LNP system formed by an ionisable lipid (DLin-MC3-DMA) had an inverse H2 structure when complexed with mRNA at low pH and this unique structure had significant influence on the transfection efficiency of the mRNA.13 Molecular dynamics simulation studies also demonstrated that RNA-LNP complexes had an inverse self-assembled structure, characterised by a non-lamellar, electron-dense internal core.14, 15 Following these studies, it is generally considered that the inverse self-assembled structures of RNA-lipid complexes play a crucial role in endosomal escape process via destabilising subcellular membranes of endosomes/lysosomes and facilitating the successful release and transfection of the RNA therapeutics.4, 12, 17 In fact, non-lamellar inverse structures formed by lipid self-assembly have attracted extensive research interests over the past few years18-21 and the advantages of the inverse structures such as the bicontinuous cubic (Q2) and the H2 phases over lamellar structures have been recognised from formulation to cellular interactions.22, 23 Therefore, understanding the LNP self-assembled structures governed by the curvature of the membrane and the bulk medium is important for the application of LNP-based delivery technology.
Few studies have examined the self-assembly of ionisable lipids on their own and the effect of pH on the final structures, let alone the kinetics of structural formation. pH is a key factor determining the structures of ionisable lipid self-assembly through ionisation of the lipid headgroup and can vary significantly in biological environments, from neutral to acidic pH 4–6 in endosomes.17 The self-assembly structures of the clinically used ionisable lipids in LNP delivery systems, to our knowledge, have not yet been investigated at their functional pH.
In this study, we first made self-assembled ionisable lipid nanoparticles (ILNPs) from two mainstream COVID-19 vaccine ionisable lipids (ALC-0315 and SM-102). Then we found a rich polymorphism of pH-dependent inverse structures for these lipids, including the inverse micellar (L2) phase, the H2 phase, and two distinct Q2 phases (Figure 1b). With the different molecular structures of ALC-0315 and SM-102, these lipids behaved differently in their self-assembly and structural evolution. The kinetic pathway of the real-time formation of the inverse structures was also tracked in situ at a millisecond timescale using time-resolved SAXS coupled with a stopped-flow rapid mixing apparatus. Understanding how ALC-0315 and SM-102 ionisable lipids build inverse ordered structures as a function of pH, as would be encountered in endosomal acidification (Figure 1c), is critical to deciphering the endosomal escape mechanism.
Additionally, aiming to expand the toolbox of the conventional LNPs for gene delivery, optimised ILNPs with the inclusion of a novel structural helper lipid monoolein (MO) have been designed herein as nanocarriers for three model nucleic acids (NAs): polyadenylic acid (polyA) tail, EGFP mRNA (mRNA), and plasmid DNA (pDNA). The kinetic pathways of SM/MO lipid complexation with NAs were also examined, observing fast multi-lamellar onion structure formation which is correlated with the specific lipid composition, and molecular composition and weight of the NAs.
Our study reports the first evidence of sequential inverse-structure formation with tuneable membrane curvature of multi-tail ionisable lipids in response to biologically relevant pH changes, as well as the structural evolution of ionisable lipid-NA complexation. This new knowledge will help to decipher ILNP-RNA nanostructures best suited for efficient RNA delivery and transfection in gene therapy.
Results and Discussion
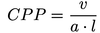 (1)
(1)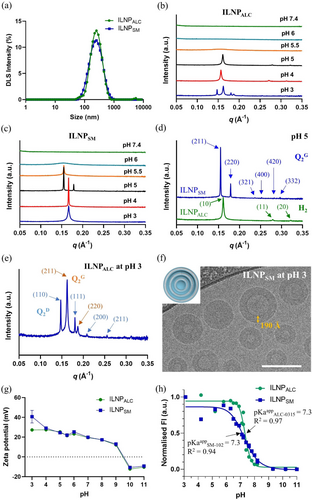
(a) DLS intensity profile of size measurement for ILNPALC and ILNPSM in water. (b,c) One-dimensional static SAXS profiles of ILNPALC (b) and ILNPSM (c) in 50 mM sodium citrate buffers pH 3–6 and sodium phosphate buffer pH 7.4. (d) Representative Bragg's peak assignment to Miller indices (hkl) of the H2 mesophase for ILNPALC (green line) and the gyroid cubic Q2G (Ia3d) mesophase for ILNPSM (blue line), both in pH 5 buffer. (e) Representative Bragg's peak assignment to Miller indices (hkl) of the double diamond Q2D (blue arrows) and gyroid Q2G (orange arrows) mesophases co-existing for ILNPALC in pH 3 buffer. (f) Cryo-TEM image of ILNPSM in pH 3 buffer, indicating a multi-lamellar onion structure (sketched in the insert). Scale bar is 100 nm. (g) pH-dependant zeta-potential, and (h) TNS assay-derived degree of ionisation measurement for ILNPALC and ILNPSM.
where v represents the effective volume of the hydrophobic tail(s), a is the effective hydrophilic headgroup area, and l is the effective length of the hydrophobic chain.18, 25 As more lipids became ionised in response to decreasing pH, the effective headgroup area (a) enlarged due to electrostatic repulsion between the charged tertiary ammonium groups, leading to a sequential structural transition towards mesophases with reduced membrane curvature, i.e. L2 to H2 to Q2, as shown in Figure 1b. In addition, even within the same mesophase, the swelling of the lattice dimensions as pH reduced was reflected by an increase in the lattice parameters (Table S1). For example, the H2 phase lattice parameter increased from 45.0 to 46.3 Å for ILNPALC as pH decreased from 5 to 4.
On the other hand, the ILNPSM system (Figure 2c) exhibited a different pH-dependent mesophase structural transition compared to ILNPALC (Figure 2b) but the same trend of decreasing membrane curvature with decreasing pH. SAXS screening of ILNPSM in different pH buffers revealed no diffraction peaks in a neutral environment (pH 7.4), and the formation of a L2 structure at pH 6, H2 structure at pH 5.5, and gyroid Q2G structure at pH 5 (mesophase determination given in Figure 2d). A single Bragg peak in the SAXS profiles of ILNPSM at pH 3 and pH 4 conditions made it difficult to assign the mesophase but following the phase transition order of decreasing membrane curvature and the former published work on the structure transformation between lamellar and cubic mesophase,26, 27 a multi-lamellar onion-like structure can be inferred. Meanwhile, the Bragg peak position (0.167 Å−1) of this onion structure at pH 3 and 4 is close to the strongest peak position (0.155 Å−1) of gyroid Q2G at pH 5, which matches the epitaxial relationship between multi-lamellar onion and cubic structures introduced in previous work.28 A correlation distance for the single peaks at pH 3 and pH 4 of 37.6 Å can be derived (Table S1). In addition, cryogenic transmission electron microscopy (cryo-TEM) of ILNPSM at pH 3 (Figure 2f) demonstrated a spherical morphology of the particles, some with hollow cores to a certain degree. The rims of the imaged nanoparticles have different thickness, which could be dictated by the number of lipid membrane layers in each particle. For example, the rim thickness of one representative ILNPSM labelled in Figure 2f is 190 Å, which corresponds to 5 lipid bilayers with the correlation distance (37.6 Å) between lipid bilayers from SAXS. We have also undertaken the cryo-TEM imaging of ILNPALC and ILNPSM at pH 5 (Figure S2) and found that the ILNPs had particle diameters of 100 to 150 nm consistent with the DLS results and internal electron dense structures. In addition, we observed some faceted particles in Figure S2, the formation of which is consistent with a nonlamellar type of symmetry. However, the inner organisation of the mesostructured lipid bilayers in the ILNPs is not well visible (Figures 2f and S2) due to the low electron density of the studied ionisable lipids.
Considering the similar amino headgroup structure of the ALC-0315 molecule and SM-102 molecule, ALC-0315 with a 4-carbon chain structure tends to form inverse structures with higher membrane curvature than SM-102 which only has 3 longer carbon chains under the same pH condition, suggesting the number of carbon chains acts more efficiently in increasing the membrane curvature than the carbon chain length. This trend can be understood according to the CPP (Equation 1) as the number of carbon chains only increases v and result in increased CPP values, but the length of carbon chain contributes to both v and l.
To gain a quantitative insight of the pH-induced phase behaviour of ILNPALC and ILNPSM, we obtained the CPP factors (v, a, and l) of individual ALC-0315 and SM-102 molecules equilibrated in an aqueous environment.15 The CPP value of neutral and charged ionisable lipids (i.e. pH effect) was then determined according to Equation 1. Specifically, for the neutral and charged ALC-0315 molecule, v is 1367 Å3 and 1564 Å3, a (the area per lipid in lipid bilayer) is about 82 Å2 and 119 Å2, respectively, and l is 12.5 Å (which is half of the bilayer thickness). The corresponding CPP value of ALC-0315 molecule is 1.33 (neutral) and 1.05 (charged). On the other hand, for the neutral and charged SM-102 molecule, v is 1347 Å3 and 1342 Å3, a is about 77 Å2 and 104 Å2, respectively, and l is 13.5 Å. The CPP value of SM-102 molecule is 1.30 (neutral) and 0.96 (charged). We can observe that the decrease of CPP value of ALC-0315 and SM-102 from a neutral status to a charged status correlates with the phase transition with reduced bilayer curvature in Figures 2b,c. In addition, as the CPP value of ALC-0315 is larger than that of SM-102, it predicts that ALC-0315 will exhibit a more curved structure than SM-102 under similar environmental conditions, consistent with our SAXS results.
Zeta-potential measurements showed a similar trend for both ILNPALC and ILNPSM changing their surface charge density in response to pH in the range of 3–11 in the bulk solution environment (Figure 2g). Both ILNPs were negatively charged (approximately −10 mV) at higher pH over 10, crossed zero around pH 9.5, and became positively charged at pH below 9. The negative zeta potentials at high pH are most likely a result of base-catalysed hydrolysis of the ionisable lipid ester bonds resulting in the formation of anionic fatty acid salts.29 The zeta potential of ILNPs were approximately 12 mV at pH 9 and further increased as pH continued to decrease, reaching 40 mV for ILNPSM and 27 mV for ILNPALC, respectively, at the acidic environment with the lowest pH 3.
Apparent pKa (pKaapp) of the ILNPs was determined by using a TNS assay.30-32 In the TNS assay, the specific structure and formulation of ILNPs are influential. Thus, the measured pKaapp reflects the population of ionisable lipids residing at the self-assembled lipid-aqueous solution interface of ILNPs rather than just the individual ionisable lipid.33
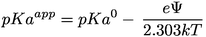 (2)
(2)where pKa0 is the intrinsic interfacial pKa of the ionisable lipid, e is the elementary electrostatic charge, Ψ is the mean field surface potential at the average site of residence for the ionisable lipid, k is the Boltzmann constant, and T is the absolute temperature.34-36 As Ψ is governed by the surface charge density of charged ionised lipids, pKaapp will therefore vary as a function of pH in the range where the lipid population exists in the uncharged state to the fully charged state.
With the pKaapp determined from the TNS assay and the assumption of the zeta-potential values of ILNPALC (30±2 mV) and ILNPSM (39±1 mV) in water as the mean field potential Ψ, the pKa0 of ionisable lipids can be calculated to be 7.8 for ALC-0315 and 8.0 for SM-102. Based on the ionisation behaviour of weak bases in organic solvent-water mixtures, a pKa0 value close to 8.0 reflects a lipid-water interfacial microenvironment with an effective dielectric constant (Ɛeff) of approximately 30.35 This Ɛeff for the ionisable lipid-water interface is comparable to that reported for the interfacial microenvironment of nonionic surfactant micelles.35
Growing evidence show that the optimal pKaapp range of nanoparticles delivering the highest siRNA activity lies in a narrow pH region between 6 and 7.8, 17 However, scarce systematic studies have been performed for mRNA-LNP delivery systems. Based on our finding, the derived intrinsic pKa0 values can accelerate the pKaapp optimisation approach by estimating pKaapp for numerous formulations without the need for performing tedious TNS assays. Finally, in vitro cell viability of ILNPALC and ILNPSM were characterised using L929 cells (Figure S4). Although with different molecular structures and pH-dependent phase behaviour, both ILNPALC and ILNPSM had a negligible toxicity effect after 24 h incubation with the cells (with a pH of about 7) up to a lipid concentration of 1600 μg/mL, which suggests good biocompatibility.
In order to understand the kinetics of the pH-driven inverse structure formation of the ionisable lipids, we performed time-resolved SAXS at the millisecond (ms) timescale at the ID02 beamline, ESRF. Rapid microfluidic mixing has been widely used for the formation of various LNPs with advanced size control for drug delivery.29, 37, 38 Combining with the in situ SAXS characterisation, the high-brilliance synchrotron X-ray source coupled with a stopped-flow rapid-mixing device at the ID02 beamline has enabled this technique to be used as a powerful tool to capture the rate of structural formation and intermediate structural evolution of biomembranes and many other self-assembled systems.28, 39-41 Capturing the inverse structure formation of ALC-0315 and SM-102 in real-time during acidification as would be encountered in endocytosis is of importance for elucidating the mechanism of endosomal escape, via inverse structural formation, in order to optimise future LNPs. In the in situ rapid mixing, ILNPs pre-made in water showing no structure under SAXS (Figure S5) were rapidly mixed with sodium citrate buffer (50 mM, pH 5) at a flow rate ratio of 1 : 1 and the SAXS profiles were recorded every 5 ms for a total of 150 ms (30 frames). pH 5 was chosen because this point showed the most prominent inverse structure formation for both lipids according to our static SAXS results (Figures 2d) and it also mimics the intracellular endosomal pH environment during the endosomal escape process.
3D and 2D time-resolved SAXS patterns during the mixing of ILNPALC or ILNPSM with the acidic buffer solution for a duration of 150 ms are presented in Figures 3a–d. We observe the kinetic pathway of the appearance of Bragg's diffraction peaks and peak area increase belonging to the different inverse structures (H2 for ILNPALC and Q2G for ILNPSM) before finally reaching a plateau within the examined timeframe. The peak position also exhibited a slight shift to the lower q during the structural evolution before reaching equilibrium indicating a growth in the lattice dimension during mixing (Figures 3b,d), but no other intermediate structures were observed at the 5 ms scanning interval. Furthermore, there is no obvious difference in the peak position and peak area observed between the last kinetic frame at 150 ms post-mixing and the SAXS pattern obtained at 15 min post-mixing (Figure S6), indicating that structural growth reached an equilibrium within 150 ms. This reveals that decreasing pH and adding ionic strength in the bulk aqueous environment induced a fast structural transformation of ILNPs into inverse ordered structures over milliseconds via ionisation of the lipid headgroup.
3D and 2D time-resolved SAXS patterns showing the kinetic pathway of the inverse structure formation of ILNPALC (a,b) and ILNPSM (c,d) pre-made in water upon in situ rapid mixing with sodium citrate buffer (50 mM, pH 5) performed at the ID02 beamline, ESRF. Schematic diagrams of the H2 phase and Q2G phase are shown as the insert in (b, d). (e) Plot of the major Bragg peak areas, A(t), as a function of time and the fitting according to Equation 3. The R2 value of the fitting is 0.92 for ILNPALC and 0.97 for ILNPSM.
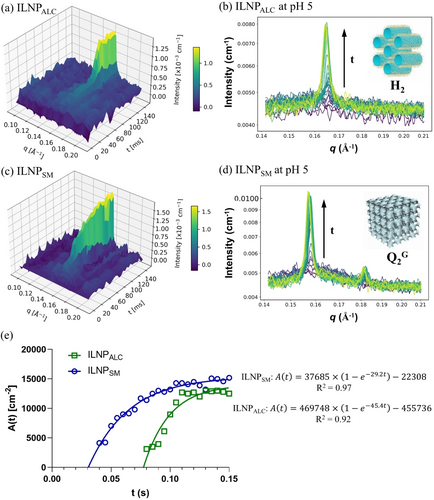
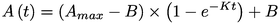 (3)
(3)where Amax is the maximal peak area, K is a rate constant analogous to a first-order chemical reaction constant, B is the intercept, and t is the time. The fitted rate constant K is 45.4 s−1 for ILNPALC with R2 of 0.92 and 29.2 s−1 for ILNPSM with R2 of 0.97, respectively. Overall, the phase transition of ILNPSM into a Q2G phase starts earlier but the structure grows slower than that of ILNPALC into a H2 phase.
The unusual ability of the ionisable lipids ALC-0315 and SM-102 to form H2 and Q2 mesophases upon acidification, as a part of their pH-dependent structural polymorphism and inverse mesophase formation with millisecond kinetics, was clearly demonstrated for the first time, with critical implications for the development of mRNA-LNP vaccines and RNA therapeutics. A sequential transition of highly ordered lyotropic liquid crystalline structures with tuneable membrane curvature (micellar to hexagonal to cubic to lamellar) can be induced by gradual changes in pH, which may result in a good endosomal escape ability following the mechanistic Scheme in Figure 1c. This is expected to contribute to the elucidation of structure–activity relationships for LNPs consisting of ionisable lipids and the development of LNP technologies with excellent endosomal escape ability, especially for intracellular gene delivery.
Despite the huge success of the COVID-19 mRNA vaccines and the promising great potential of the LNP technology for nucleic acid (NA) delivery, the structural evolution of NA complexation with ionisable lipids has been rarely reported, let alone its kinetics at a millisecond timescale. We continued to study the lipid/NA complexation-induced structural formation and kinetics. As an initial screening, pure ionisable lipids (ALC-0315 or SM-102) in an organic solvent were mixed with polyadenylic acid (polyA) tail as the model NAs in a pH 6.5 sodium citrate buffer at various ratios and static SAXS measurement revealed no diffraction peak (Figure S7a), indicating lack of ordered structures. Most reported and clinically approved ILNPs as drug delivery vehicles and nanovaccines consist of mixed components including the ionisable lipids and some helper/PEGylated lipids, with the ionisable lipids being ca. 50 mol% of the total lipids.2, 5, 13, 16 Altering the helper lipid components can change the structure formation of the final NA-loaded LNPs.12 Therefore, a helper lipid, monoolein (MO) well known to form inverse Q2 and H2 structures,18-20, 42 was added at a 1 : 1 molar ratio to the ionisable lipids, achieving ILNPALC/MO and ILNPSM/MO.
We first characterised the pH-dependent mesophase formation, morphology, size stability and pKaapp of ILNPALC/MO and ILNPSM/MO (Figure 4) to determine their property differences to the ILNPs of pure ionisable lipids. MO is an inverse structure-forming lipid (Figure 4a), self-assembling into an internal primitive cubic Q2P phase when dispersed into nanoparticles with Pluronic F127 polymers.18-20, 42 Figure 4b shows both ILNPs are stable at room temperature as there is no obvious size change. Both ILNPALC/MO and ILNPSM/MO in buffer solution (10 mM, pH 5) were further characterised to confirm their nanoparticle morphology and size, using cryo-TEM (Figures 4c,d). For phase information, static SAXS (Figures 4e,f) of ILNPALC/MO and ILNPSM/MO in different pH buffers revealed no diffraction peak at the neutral environment (pH 7.4). With pH decreasing, ILNPALC/MO formed an L2 structure at pH 6 and H2 structure from pH 5.5 to 3 with a lattice parameter increasing from 42.5 Å to 48.2 Å. On the other hand, ILNPSM/MO transformed into a mixed L2/H2 structure at pH 6, H2 structure at pH 5.5 and 5, and gyroid Q2G structure at pH 4 and 3. The phase transition of both ILNPs as the pH decreased followed the order of mesophases with decreasing membrane curvature. Compared with the phase behaviour of the ILNPALC and ILNPSM at the corresponding pH in Figures 2b,c, the addition of the inverse structure-forming lipid MO increased the membrane curvature and extended the pH range whereby inverse structures were observed. This demonstrates the feasibility to control the specific pH-dependent phase behaviour of ALC/SM-based nanosystems by adjusting the ionisable lipid/helper lipid (in this case MO) molar ratio.
(a) Molecular structure of MO and a schematic drawing of primitive cubic Q2P phase. (b) DLS intensity profiles of ILNPALC/MO and ILNPSM/MO before and after 14-day storage at room temperature. (c,d) Cryo-TEM image of ILNPALC/MO (c) and ILNPSM/MO (d) in citrate buffer solution (10 mM, pH 5). Scale bar: 100 nm. (e,f) One-dimensional static SAXS profiles of ILNPALC/MO (e) and ILNPSM/MO (f) in 50 mM sodium citrate buffers with pH from 3–7.4. (g) TNS assay-derived degree of ionisation measurement for ILNPALC/MO and ILNPSM/MO. (h) The values of pKaapp at 50 % ionisation measured using TNS assay and calculated according to Equation 2.
The pKaapp of ILNPALC/MO and ILNPSM/MO was measured to be 7.6 and 7.5 (Figure 4g). As MO is a neutral lipid in the system,43 the zeta potential of ILNPALC/MO and ILNPSM/MO in H2O was measured to be 40±1 mV and 38±1 mV, which is closer to that of the ILNPs without MO (30±2 mV for ILNPALC and 39±1 mV for ILNPsM). As comparison in Figure 4h, using the zeta potential as Ψ and calculated pKa0 (7.8 for ALC-0315 and 8.0 for SM-102), the pKaapp of ILNPALC/MO and ILNPSM/MO was calculated to be 7.1 and 7.4 according to Equation 2, similar with the corresponding experimental value (7.6 and 7.5) obtained from the TNS assay, which supports the feasibility of this fast first-order determination of pKaapp for ionisable lipid-based ILNPs containing ALC-0315 and SM-102.
A number of studies have screened large nanoparticle materials libraries elucidating relationships between physical properties of the nanoparticle systems and the transfection/gene silencing activities of the delivered RNAs.8, 33 In our lipid-nucleic acid complexation (NA@lipid) study, we first observe an interesting phenomenon that the polyA@lipids complexation process in citrate buffer (1 mM, pH 6.5) can induce a multi-lamellar onion structure indicated by the appearance of a sharp SAXS peak at q=0.147 Å−1 for the SM/MO formulation, but no peaks observed for ALC/MO formulation (Figure S7b). As we discussed before, ILNPSM/MO had a mixed L2/H2 structure at pH 6 and no peak observed at pH 7.4 (Figure 4f). However, the pH-dependent electrostatic binding between the ionisable cationic SM-102 and the negatively charged polyA will significantly change the distribution of ionisable lipids in individual ILNP and the self-assembled structures.44 For our case, the complexation of polyA at pH 6.5 decreases the curvature of the lipid membrane and induces the formation of the multi-lamellar onion structure of polyA@SM/MO (Figure S7b). Meanwhile, as the polyA amount increases from 5 wt% to 20 wt% relative to the total lipid mass, corresponding to a molar ratio between nitrogen in the ionisable cationic lipid population and phosphate in the nucleotide population (N/P molar ratio)5 from 1.2 to 4.8, there is neither phase transition nor peak position shift observed (Figure S7b). This suggests that the mesostructure of polyA@SM/MO is independent from the N/P molar ratio in a range of 1.2 to 4.8, which may be contributed to the excess lipid amount to polyA. On the other hand, the reason for no ordered structure being observed in polyA@ALC/MO is not clear (Figure S7b). Therefore, due to the lack of mesostructure observed in ALC/MO formulations, we focused on using SM/MO formulations in this kinetic study.
Due to the impossibility of removing organic solvent during the in situ real-time SAXS measurement, the effect of adding stabiliser and dialysis (solvent removal) on the final structure of NA@SM/MO was investigated using polyA and GFP mRNA (Figures S8a,b). The DLS data show that in the absence of a stabiliser, massive clustering and fusion into large NA@lipid complexes existed during the dialysis. Adding the steric stabiliser (Pluronic F127) can hamper the aggregation process leading to more homogeneous and smaller size of the NA@lipid complexes, especially after the dialysis. However, the structure formation as seen in the SAXS study seems not to be affected by stabiliser nor dialysis. Figures S8c,d show a single Bragg peak in the q of 0.147 Å−1 (correlation distance=42.7 Å) and the position of the peak did not change when F127 was added and after dialysis.
For the kinetic study at the ID02 beamline, three types of NAs were employed, including a polyA (single-stranded, 2100—10000 nt, Mw 700–3500 kDa), a GFP mRNA (single-stranded, 996 nt, Mw 320 kDa) and a plasmid DNA (pDNA, double-stranded, 5487 bp, Mw 3390 kDa). As sketched in Figure 5a, the in situ rapid-mixing apparatus mounted at the ID02 beamline has the solvent solution and aqueous solution simultaneously injected into the microfluidic channel with a fixed 1 : 3 volume ratio and a total flow rate of 6.7 mL/s, which is comparable with the manufacturing conditions (over 20 L/h) of both Pfizer and Moderna COVID-19 mRNA vaccines using NanoAssemblr GMP System.29 In 3D time-resolved SAXS patterns (Figures 5b–d), the appearance and growth of the single peak were observed between 0.12 Å−1 and 0.15 Å−1 for all the complexation during the recording time of 150 ms. The last SAXS profiles of the three NA@SM/MO complexation processes acquired at 150 ms are plotted in Figure 5e, which shows the three different classes of NAs can direct self-assembly into ordered mesostructures with various dimensions. Specifically, among the three NAs, polyA@SM/MO demonstrated a sharp peak which may be attributed to its simple molecular structure (only adenine bases). The peak appears at the largest q position (0.146 Å−1), revealing a multi-lamellar onion structure with the smallest lattice parameter of 43 Å. This suggests a strong interaction between adenine bases and lipids.
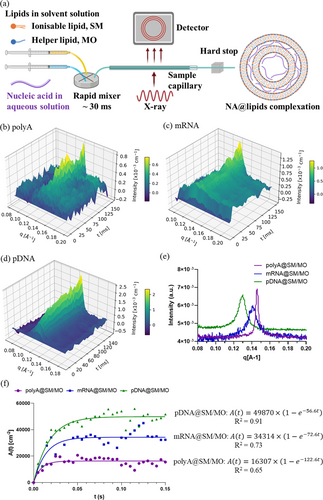
(a) Schematic presentation of the principle of the millisecond time-resolved SAXS for the structural measurement during NA@lipids complexation, performed at the ID02 beamline, ESRF. (b–d) 3D time-resolved SAXS patterns showing the kinetic pathway of the self-assembled process of polyA (b), mRNA (c), and pDNA (d) complexation with SM/MO lipids. (e) SAXS profiles of all three complexes in the final state taken at 150 ms post-mixing. (f) Plot of the major Bragg peak areas, A(t), as a function of time during the complexation of NAs (PolyA, mRNA, and pDNA) with lipids (SM/MO) and the fitting according to Equation 3 with the intercept B set to be 0.
A single peak pattern was also observed at 0.141 Å−1 for mRNA@SM/MO (Figure 5c) and at 0.130 Å−1 for pDNA@SM/MO (Figure 5d), showing a consistent multi-lamellar onion structure with PolyA@SM/MO. This finding also matches the reported behaviours of mRNA@lipid complex and DNA@lipid complex, both favouring a multi-lamellar structure with zero bilayer curvature.45 The peaks of mRNA@SM/MO and pDNA@SM/MO are broader than that of polyA@SM/MO, which can be attributed to more complicated molecular compositions based on four nitrogenous bases (adenine, guanine, cytosine, and uracil) of mRNA and pDNA. The peak position at 0.130 Å−1 of pDNA@SM/MO is in a good agreement with that (0.125 Å−1) of pDNA@MO/DOMA/DOPE-PEG2000 lipoplex particles reported in the former work.40, 46 The lattice parameter of pDNA@SM/MO was 48 Å, which is larger than mRNA@SM/MO (45 Å). This reveals the space between the lipid bilayers of pDNA@SM/MO is larger than mRNA because of a double-stranded structure and a larger molecular weight of pDNA. The single-peak SAXS profile of the NA@SM/MO is in an agreement with that of the mRNA-loaded DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 LNPs reported in the literature.13 Meanwhile, it has also been reported that the polyA@DLin-MC3-DMA:DSPC:Chol:DMPE-PEG2000 in bulk phase demonstrated a H2 phase at an acidic environment (pH 3) and a single-peak SAXS profile in a neutral environment,13 which may be a possible phase transition situation for our NA@SM/MO during the endosomal escape process. The pH-dependent phase behaviour and the biological performance of the NA@SM/MO in correlation with the structures is still under investigation and is not the focus of this work. The structural variation of NA@SM/MO with the addition of other components,2, 5, 12, 47, 48 such as helper lipids (e.g. DOPE, cholesterol), co-surfactant (e.g. hexanol), or stabilisers (e.g. PEG-lipid or surfactant), and the adsorption of proteins49, 50 (e.g. Apolipoprotein E) is planned to be systematically studied in future work.
The time dependence of peak area A(t) of the three NA@SM/MO demonstrates the kinetic profiles of the complexation process (Figure 5f). Practically, lipid supersaturation will be produced and electrostatic coupling between cationic lipids and negatively charged NAs will quickly form when the lipid solution is combined with an aqueous solution containing NAs. This promotes the direct self-assembly of nanostructured NAs@SM/MO complexes. As shown in Figure 5f, when the time-dependent A(t) of NA@SM/MO was fitted using Equation 3 with the intercept B=0, we found the rate constants K for polyA@SM/MO, mRNA@SM/MO, and pDNA@SM/MO, as 122.6, 72.6, and 56.6 s−1. We observe that the rate constant of the structure formation during the NA@SM/MO complexation depends on the specific NA molecule. The rate of polyA@SM/MO (K=122.6 s−1) is the fastest, which may be attributed to the simpler molecular structure of polyA. For mRNA and pDNA, K (72.6 s−1) of mRNA@SM/MO is faster than that (56.6 s−1) of pDNA@SM/MO, which may manifest from the smaller molecular weight and single-stranded molecular structure of mRNA. In addition, the rate constant K (56.6 s−1) of this direct self-assembled pDNA@SM/MO is greater than the previously reported fastest K (31.4 s−1) for the complexation of pDNA with nanostructured lipid nanocarriers,40 accelerated by the transient lipid oversaturation created during solvent mixing.29 The millisecond kinetic of nucleic acid-lipid complexation under a comparable flow condition with the manufacturing of COVID-19 vaccines reveals the real rates of the formation of self-assembled structures between various classes of nucleic acids and ionisable lipids. This knowledge can guide future LNP-nucleic acid formulation optimisation efforts, enhance our understanding of the factors influencing LNP performance, and facilitate the development of more efficient delivery systems for mRNA and gene therapies.
Conclusion
By studying the kinetics of complexation of LNPs and mRNA by time-resolved SAXS and cryo-TEM, further insights are gained into how LNPs can encapsulate and protect mRNA from degradation and how they can release mRNA into the cytoplasm. These insights can help the development of new vaccines against various diseases by improving the safety, stability, potency, and efficacy of mRNA delivery. In summary, we have demonstrated that the widely used ionisable lipids ALC-0315 and SM-102 form many inverse mesophases in acidified medium as a part of their diverse mesophase formation and pH-dependent structural polymorphism, with quantitative insight according to CPP equation. For the first time, the millisecond kinetics of the unusual ability of the ionisable lipids ALC-0315 and SM-102 to form H2 and Q2 mesophases upon acidification (pH 5) was investigated by in situ high resolution time-resolved SAXS coupled with rapid flow mixing, revealing a fast rate constant of 45.4 s−1 for ILNPALC and 29.2 s−1 for ILNPSM, respectively. The pH range of ILNPs, whereby inverse structures were observed, was further extended with the inclusion of 50 mol% MO as a novel inverse structure-forming helper lipid. The helper lipid MO also provides an environment which can influence the pKa, the kinetics of the assembly pathway of the ILNPs, and their stability. The evolution of the multi-lamellar onion structure formation upon ionisable-lipid-RNA/DNA complexation are also quantitatively revealed, showing a super-fast and RNA/DNA-dependent formation kinetic with rate constant of 122.6 s−1 for polyA, 72.6 s−1 for mRNA, and 56.6 s−1 for pDNA. The new knowledge gained from the performed millisecond kinetic study of nucleic acid-lipid complexation is expected to stimulate further developments in the field of mRNA-LNP vaccines, delivery of genetic material, and RNA-ILNP therapeutics.
Supporting Information
The authors have cited additional references within the Supporting Information.51-56
Acknowledgments
JZ gratefully acknowledges the financial support from a 2020 VESKI Victoria Fellowship (BMIN-2-20-7044) by the Victorian Government, Australia. JZ and CJD acknowledge funding support from the mRNA Victoria Research Acceleration Fund Round 1 (BMIN-2-22-19942). JZ also acknowledges travel funding support from the Australian Synchrotron International Synchrotron Access Program Round ISAP 2022/2 (ISP18998). BA obtained financial support from the projects ‘‘Structural Dynamics of Biomolecular Systems” (ELI-BIO) (CZ.02.1.01/0.0/0.0/15_003/0000447) and ‘‘Advanced research using high-intensity laser produced photons and particles” (CZ.02. 1.01/0.0/0.0/16_019/0000789) from the European Regional Development Fund. AA acknowledges a membership in CNRS GDR2088 BIOMIM research network. BD acknowledges support from the Australian Research Council Discovery Early Researcher Award (DE230101504). We thank the ESRF facility and their staff for all the support provided during our merit beam time (SC-5327) at the ID02 beamline. We also thank the ANSTO Australian Synchrotron (AS) SAXS/WAXS beamline staff, in particular Dr Susi Seibt and Dr Nigel Kirby, for their support during our experiments at the AS beamline. Finally, we thank the Micro Nano Research Facility (MNRF) at RMIT University for the use of the PC2 facility. Open Access publishing facilitated by RMIT University, as part of the Wiley - RMIT University agreement via the Council of Australian University Librarians.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.