Loading of Amino Acids onto RNA in a Putative RNA-Peptide World
Graphical Abstract
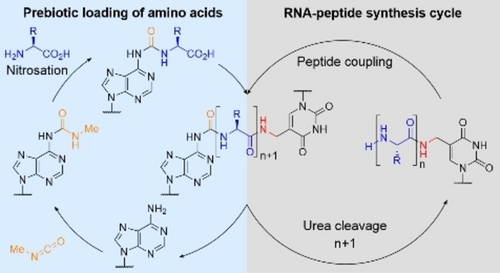
The loading of RNAs with amino acids constitutes the first step toward RNA-based peptide synthesis in a putative prebiotic RNA-peptide world. The efficient loading of RNA strands incorporating N-methylcarbamoyl nucleotides with a series of amino acids is reported. The loaded RNA strands also underwent RNA-based amino acid transfer reactions.
Abstract
RNA is a molecule that can both store genetic information and perform catalytic reactions. This observed dualism places RNA into the limelight of concepts about the origin of life. The RNA world concept argues that life started from self-replicating RNA molecules, which evolved toward increasingly complex structures. Recently, we demonstrated that RNA, with the help of conserved non-canonical nucleosides, which are also putative relics of an early RNA world, had the ability to grow peptides covalently connected to RNA nucleobases, creating RNA-peptide chimeras. It is conceivable that such molecules, which combined the information-coding properties of RNA with the catalytic potential of amino acid side chains, were once the structures from which life emerged. Herein, we report prebiotic chemistry that enabled the loading of both nucleosides and RNAs with amino acids as the first step toward RNA-based peptide synthesis in a putative RNA-peptide world.
Introduction
The essential biological process of translation converts a genotype into a phenotype. It is based on information-encoding messenger RNA (mRNA), the ribosome as the catalytic entity, and transfer RNAs (tRNAs) that carry specific amino acids and bind to the information-encoding units on the mRNA.1, 2 The attachment of an specific amino acid to its cognate tRNA (aminoacylation) requires the activation of the amino acid as an adenylate3 and a dedicated aminoacyl tRNA synthetase,4-6 which transfers the amino acid to the 3′-CCA end of the tRNA by catalyzing the formation of an ester bond. Although tRNAs are considered to be highly conserved structures,7-10 the emergence of this complex process is still one of the main unsolved questions in origin of life research.
The main problem for chemical evolution is that complex chemical structures, such as mRNAs, ribosomes and amino acid loaded tRNAs, are required to generate a machinery that can efficiently form peptide bonds. But how could such complex systems have evolved in the absence of efficient peptide synthesis machines? To solve this chicken-and-egg conundrum, we recently postulated that RNA-amino acid conjugates could have been a starting point for evolving more complex systems.11 We learned that RNA was capable of self-decorating with peptides with the help of non-canonical nucleosides,12 that is, N6-methylated derivatives of glycine- and threonine-modified N6-carbamoyl adenosine, 1 a (g6A)13 and 1 b (t6A).14 In addition, 1 a–b are found in contemporary tRNAs in all three kingdoms of life15, 16 as potential relics of an early RNA world.17, 18 In aqueous solution, these compounds exhibited a good stability over a wide pH range at moderate temperatures due to the urea bond.19, 20 In the presence of a second non-canonical nucleoside, namely, 5-methylaminomethyl uridine (mnm5U),21, 22 and when both nucleosides were placed in close proximity by RNA hybridization,23 we observed the formation of peptides. This RNA-templated reaction established a primitive peptide synthesis cycle that afforded RNAs decorated with peptides. Although our system is far away from a translational machinery, it showed that RNA-peptide chimeras, having, in principle, information-encoding properties and an expanded repertoire of functional groups, could have existed in a prebiotic world.
We would like to emphasize that the obtained RNA-peptide chimeras are not considered by us to be the direct precursors of the contemporary ribosome. In this regard, seminal works by other research groups showed that amino acids could be connected to 5′-phosphate groups as acyl phosphate mixed anhydrides24 or phosphoramidates,25-31 and transferred to the ribose 2′/3′-hydroxy group32 of terminal nucleotides in RNAs.33-40
The first step of the described peptide growth on RNA, however, required the efficient loading of amino acids onto N6-carbamoyl adenosine nucleotides. Previously, we reported that amino acids, namely, Gly and Thr, could react with methylisocyanate (MIC) affording N-methylcarbamoyl amino acids in quantitative yields (Scheme 1a).41 After nitrosation of the N-methylcarbamoyl amino acids, the obtained N-nitroso derivatives decomposed at pH 8 and 70 °C to give N-isocyanates, which reacted with adenosine to provide the required amino acid modified N6-carbamoyl (1 a and 1 b) and O-carbamoyl nucleosides. While this chemistry rested on prebiotically plausible starting materials, it had the drawback that the yields were sensitive to the exact reaction conditions and fairly low.
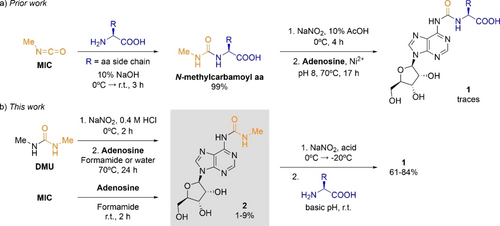
Prebiotic synthesis of amino acid modified N6-carbamoyl adenosine nucleosides (1) from: a) N-methylcarbamoyl amino acids and b) N6-methylcarbamoyl adenosine (2). aa=amino acid.
Herein, we show that a prebiotically more plausible loading of amino acids is possible with N6-methylcarbamoyl adenosine 2 (Scheme 1b). The loading reaction was so efficient that it was also possible when 2 was embedded in RNA strands. The reported chemistry could be expanded to N2-methylcarbamoyl guanosine 3 and N4-methylcarbamoyl cytidine 4. The obtained results suggest that the loading of amino acids onto RNAs constitutes an additional property, along with information-encoding, which needs to be considered in the context of prebiotic chemistry.
Results and Discussion
Prebiotically Plausible Synthesis of N6-Methylcarbamoyl Adenosine
Initially, we investigated the formation of the urea-modified adenosine 2 from either 1,3-dimethylurea (DMU) or methylisocyanate (MIC) under prebiotically plausible reaction conditions (Scheme 1b). Both DMU and MIC are prebiotically plausible starting materials. MIC could be generated from CH4 and HNCO by vacuum-UV light irradiation at 20 K.42 Moreover, MIC was detected in the comet 67P/Churyumov-Gerasimenko43 and in the protostar IRAS 16293-2422.42, 44 In turn, DMU was formed by continuous flow plasma discharge experiments from gas mixtures (N2/CO/CO2/H2).45 It is also possible that DMU was produced by the combination of MIC with methylamine, which was also present on the above-mentioned comet.43
In a 0.4 M HCl aqueous solution at 0 °C, the reaction of DMU with 1 equiv of NaNO246-48 gave its N-nitroso derivative in 98 % yield.49 Subsequently, the addition of adenosine to a formamide43, 50 or an aqueous solution, containing 50 equiv of the N-nitroso derivative of DMU, followed by heating at 70 °C afforded the N6-carbamoylated adenosine 2 in up to 2 % yield. The high-performance liquid chromatography—mass spectrometry (HPLC-MS) analyses of the crude reaction mixtures showed the formation of 2, as well as other adenosine derivatives having either methyl or N-methylcarbamoyl substituents, or both (Figure S1a–b). Most likely, the isomeric products of 2 had the N-methylcarbamoyl substituent at one of the ribose hydroxy groups, that is, O-methylcarbamoyl adenosine derivatives. In formamide solution at room temperature (r.t.), the reaction of adenosine with 5.5 equiv of MIC yielded 2 in a maximum amount of 9 % (Figure S2). Taken together, these results showed that two prebiotically plausible synthetic pathways could provide the urea-modified adenosine 2.
Next, we probed the stability of the N6-methylcarbamoyl adenosine 2 at 70 °C in aqueous solution at different pH values within the range 6–9.5. Under all these conditions, the HPLC-MS analyses, after 24 h, indicated that 2 was stable in aqueous solution. Thus, heating at 70 °C the above obtained crude reaction mixtures for 24 h in 50 mM borate buffer pH 9.5 led to the hydrolysis of some of the O-methylcarbamoyl adenosine derivatives, whereas 2 persisted in the aqueous solution (Figure S3).
Prebiotic Loading of Amino Acids onto Nucleosides
Based on our previous study on the prebiotic synthesis of amino acid modified N6-carbamoyl adenosine 1 from N-methylcarbamoyl amino acids (Scheme 1a),41 we envisaged that the analogous nitrosation of the urea-modified adenosine 2, followed by its conversion into the corresponding N6-isocyanate could also lead to the formation of 1 (Scheme 1b). First, we optimized the conditions for the reaction of 1 with Gly using HPLC-MS analyses (Table 1 and Tables S1–S4). In the first reaction step, we performed the nitrosation of 2 with 12.5 equiv of NaNO2 in an acidic aqueous solution. Among the acids tested, 5 % H3PO451 in water gave the best results, indicating that the formation of the N-nitroso derivative of 2 required rather harsh acidic conditions (pH<1.5). Next, we investigated the reaction under freezing conditions, and we found that the yield strongly improved at −20 °C for 22 h. Most likely, under these conditions, the reactants were excluded from the growing ice crystals and concentrated in the interstitial (eutectic) phase.52-55 In the second reaction step, the thawed aqueous solution, containing initially 2 and NaNO2, was adjusted to near neutral or basic pH. At r.t. and in the presence of 10 equiv of Gly, the loading of the amino acid onto 2 was very efficient at pH 9.5,56 affording 1 a (g6A) in a remarkable 84 % yield. When the amino acid was added in the first step, the nitrosation reaction, the amino acid modified N6-carbamoyl adenosine was also obtained, yet in lower yield (Figure S6 and Table S5). Note that the prebiotic formation of 1 a required dramatic changes in the pH of the aqueous solution. In this respect, it is worth mentioning that the second reaction step, coupling of the amino acid, also occurred under slightly acidic conditions in 5 % yield (Table 1). We also attempted the loading of Gly using the N6-methylated version of 2. However, under the optimized reaction conditions, the yield of the expected nucleoside product (m6g6A) was reduced to 8 % (Figure S9).
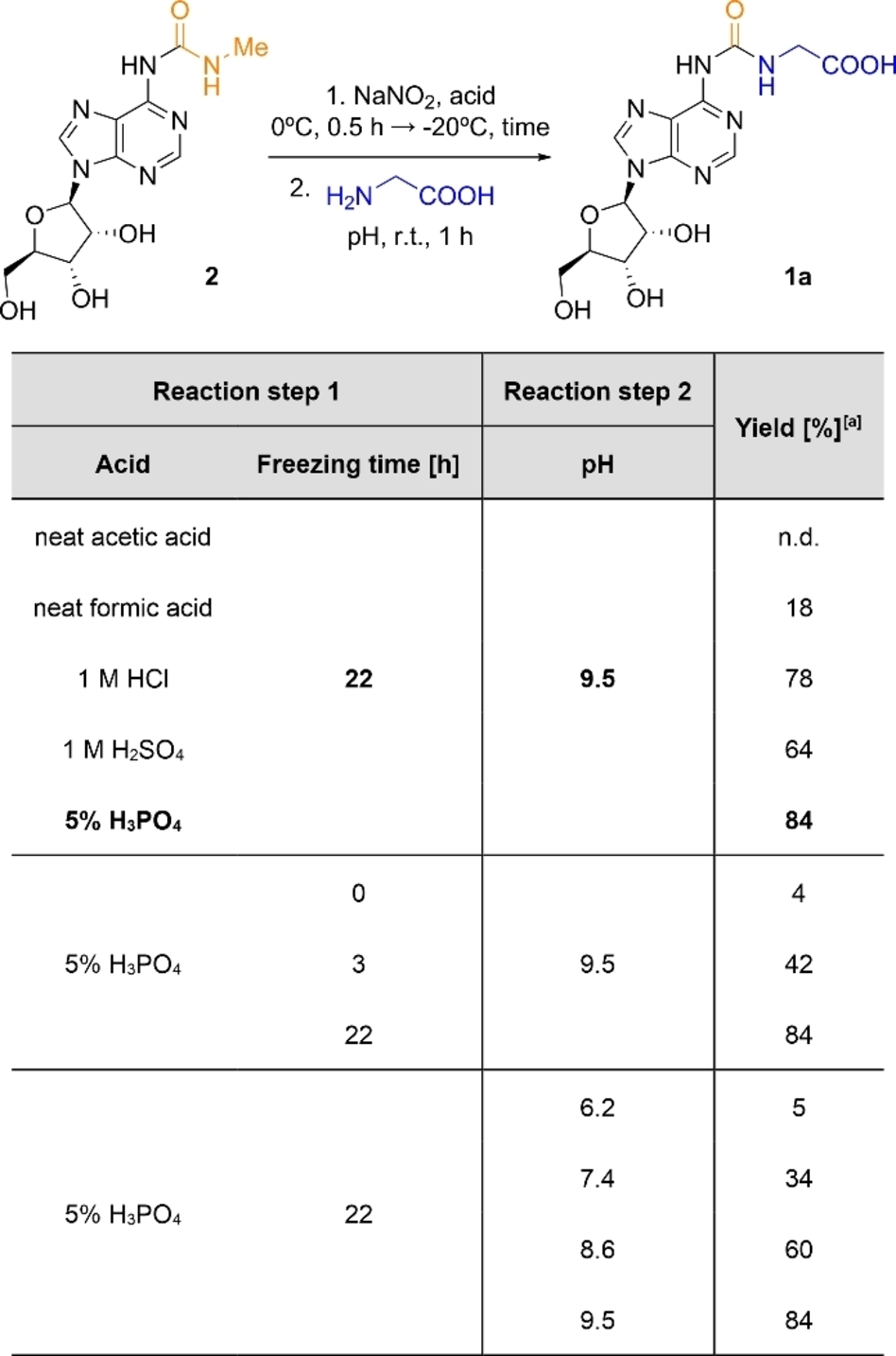
- [a] Yields determined by HPLC analysis using the calibration curve of 1 a (Figure S10). n.d.=not detected.
To evaluate the scope of the prebiotic synthesis of amino acid modified N6-carbamoyl adenosine nucleosides 1, we carried out the reaction with a series of L-amino acids, having aliphatic (Ala, Pro), aromatic (Phe), neutral polar (Thr) and ionizable (Asp, Lys) side chains (Table 2).57 Under the reaction conditions optimized above for 1 a (g6A), we observed, in all cases, the formation of the nucleoside products 1 b–g in yields between 61–78 % (Figures S4–S5). We also detected, in all cases, the presence of ca. 10 % adenosine and the remaining starting material 2 (Figure 1a). Interestingly, the reaction of 2 with Lys, with its α- and ϵ-amino groups, provided a selectivity of 80 : 20 for the product α-1 f (k6A) versus ϵ-1 f.

- [a] Yields determined by HPLC analysis using the calibration curves of reference compounds (Figures S10 and S19). [b] Reactions performed in the presence of GdmCl. [c] The structure of the Gly derivative contained a nitrile group instead of a carboxylic acid. A=adenosine. n.d.=not detected.
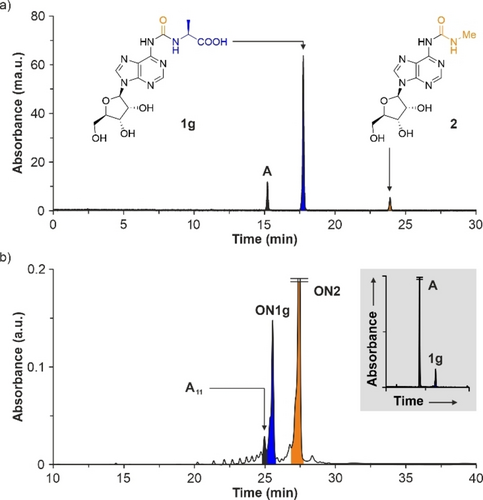
HPLC chromatograms of the crude reaction mixtures of Ala with: a) 2 and b) ON2. In b, inset shows enzymatic digestion of ON1g. A=adenosine and A11=5′-AAAAAAAAAAA-3′.
Next, we studied the loading of an amino nitrile onto N6-methylcarbamoyl adenosine 2. Amino nitriles were suggested as prebiotically plausible precursors for peptides.11, 58-61 Indeed, the reaction of 2 with the nitrile derivative of Gly, that is, GlyCN, generated the nucleoside product 1 h (gCN6A) in 26 % yield (Table 2).
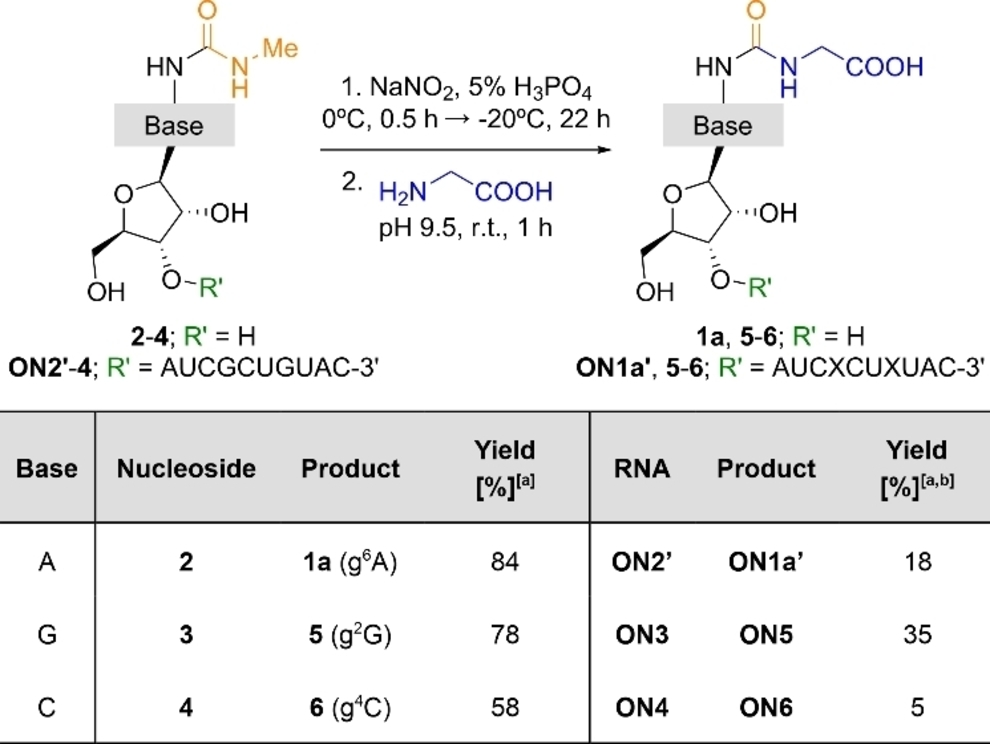
Encouraged by the reaction of 2 with amino acids, we explored the loading of Gly with other urea-modified nucleosides: N2-methylcarbamoyl guanosine 3 and N4-methylcarbamoyl cytidine 4 (Table 3). We found that the addition of Gly to an aqueous solution of 3 at pH 9.5, after its nitrosation, afforded 5 (g2G) in 78 % yield (Figure 2a). Similarly, the combination of 4 with Gly gave 6 (g4C) in 58 % yield (Figure S8). These results suggested that the putative RNA-peptide synthesis might not be limited to RNAs containing the amino acid modified N6-carbamoyl adenosine 1, although the non-canonical nucleosides 5 (g2G) and 6 (g4C) were so far not found in contemporary RNAs.62
- [a] Yields determined by HPLC analysis using the calibration curves of reference compounds (Figures S10, S11 and S19). [b] Reactions performed in the presence of GdmCl. A=adenosine; C=cytidine, G=guanosine, U=uridine and X=xanthosine.
HPLC chromatograms of the crude reaction mixtures of Gly with: a) 3 and b) ON3. In b, inset shows enzymatic digestion of ON5. A=adenosine; C=cytidine; G=guanosine; U=uridine; X=xanthosine and B11=5′-GAUCXCUXUAC-3′.
All together, these data showed that N-methylcarbamoyl nucleosides, 2–4, could be efficiently loaded with amino acids and amino nitriles in aqueous solution under prebiotically plausible conditions to form amino acid modified N-carbamoyl nucleosides.
Prebiotic Loading of Amino Acids onto RNA
We investigated whether the loading of amino acids was possible when N6-methylcarbamoyl adenosine 2 was incorporated into RNA. For these experiments, and the subsequent RNA-based amino acid transfer, we synthesized the phosphoramidite derivative of 2 in four synthetic steps with a 35 % overall yield (Scheme S19). Then, we used solid-phase RNA synthesis to incorporate the urea-modified nucleoside at the 5′-end of the 11-mer homo-A RNA strand ON2 (Table 2).
We started with the homo-A RNA strand ON2 and analyzed the loading of a series of amino acids. For example, when we treated ON2 with 500 equiv of NaNO2 in 5 % H3PO4 at −20 °C and then, we added, at r.t., an excess of Ala, followed by basification to pH 9.5, the HPLC chromatogram of the crude reaction mixture, as well as the matrix-assisted laser desorption/ionization—time-of-flight (MALDI-TOF) mass spectrum of the purified compound revealed the formation of the RNA strand ON1g with a terminal a6A nucleotide (Figure S14). Next to the remaining starting material, we noted the formation of three major side products. Using MALDI-TOF MS, we assigned the degradation products to 10-mer and 11-mer RNA strands, A10 and A11, that had lost the terminal urea-modified nucleoside and the N6-methylcarbamoyl substituent, respectively. We also detected a 10-mer RNA strand with a terminal a6A nucleotide, A9-a6A. It seems that the nitrosation reaction provoked the partial degradation of the RNA strands. However, the addition of 100 mM of guanidinium chloride (GdmCl) salt reduced the side reactions dramatically (Figure 1b), potentially by formation of salt-bridges between the (nitrosated)63, 64 guanidinium cation and the phosphodiester backbone.65 Other salts, such as NaCl and NaClO4, had little effect on the RNA degradation (Figure S14).
In order to further prove the loading of Ala onto the RNA strand ON2, we performed an enzymatic digestion of the isolated RNA product ON1g into the nucleosides, and analyzed the digest with the help of the corresponding nucleoside reference compounds by HPLC-MS.66 We treated ON1g with a nucleoside digestion mixture at 37 °C for 2 h. Analysis of the obtained nucleoside mixture showed two peaks corresponding to 1 g (a6A) and adenosine in a ca. 1 : 10 ratio (Figure 1b), which was in agreement with the nucleotide composition of the digested RNA strand.
In the presence of GdmCl and for all the amino acids studied, we observed the formation of the RNA strands ON1a–g in 10–23 % yield (Table 2 and Figures S15–S16). It is worth mentioning that the reaction of Lys with the RNA strand ON2 yielded predominantly the α-regioisomer, α-ON1f, of the k6A nucleotide. For the loading with the amino nitrile GlyCN, we did not detect the expected RNA product ON1h.
In analogy to the loading of amino acids onto different nucleosides, we evaluated the loading of Gly with three 11-mer RNA strands, ON2’–ON4 (Table 3), having all four canonical nucleotides, but bearing distinct N-methylcarbamoyl nucleotides at the 5′-end. The RNA strands were synthesized from the corresponding phosphoramidite derivatives of 2–4 (Schemes S19–S21). In all cases, the reactions of the three RNA strands with Gly, under the conditions described above with GdmCl, gave the respective products, ON1a’, ON5–ON6, in 5–35 % yield (Table 3, Figure 2b and Figure S12). MALDI-TOF MS analyses of the obtained RNA products showed the successful loading of Gly onto the parent RNA strands. In addition, we detected, after the enzymatic digestion of the RNA products, the partial conversion of guanosine to xanthosine by nitrosative deamination.67, 68
Moreover, we investigated the loading of Phe onto an RNA strand ON2’’ bearing two N6-methylcarbamoyl adenosine nucleotides, placed at the 5′-end and an internal position of the sequence (Scheme S8). Using similar reaction conditions to those described above, we observed the formation of the double-loaded RNA strand ON1c’ as the major species in the crude reaction mixture (Figure S13).
Collectively, these data showed that RNA strands containing N-methylcarbamoyl nucleotides at either terminal or internal positions, or both, could be loaded with amino acids in a sufficient extent to perform RNA-based amino acid transfer (see below).
RNA-Based Amino Acid Transfer Following the Loading Reaction
Finally, we wanted to demonstrate that the prebiotic loading of an amino acid onto an RNA strand, functionalized with a urea-modified adenosine nucleotide at the 5′-end, could undergo RNA-based amino acid transfer (Figure 3a). The loading reaction provides a terminal amino acid modified N6-carbamoyl adenosine (donor) as the basis for the synthesis reaction. For the amino acid transfer, we hybridize the obtained donor RNA strand with a complementary acceptor RNA strand, containing a 5-aminomethyl uridine at the 3′-end, forming a duplex. Activation of the carboxylic acid in the donor RNA strand leads to the formation of a hairpin intermediate, which is opened at elevated temperatures by urea cleavage, transferring the loaded amino acid to the acceptor RNA strand. The obtained acceptor RNA strand can be involved in subsequent reactions, enabling peptide growth on RNA.11 Since the nitrosation reaction converts guanosine to xanthosine, we employed a binary code of adenosine and uridine nucleotides in the initial RNA strand ON7. We also selected Phe for the experiment because of its high reaction efficiency (Table 2).11
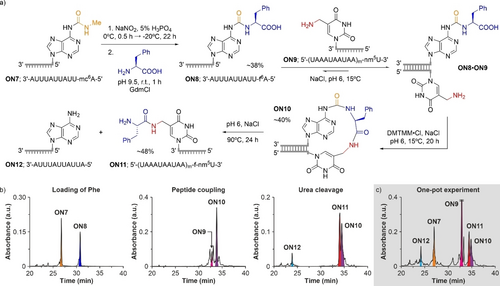
a) Reaction scheme for RNA-based amino acid transfer: loading of Phe, peptide coupling and urea cleavage. HPLC chromatograms of the crude reaction mixtures of: b) the individual steps and c) the one-pot experiment. mc6A=N6-methylcarbamoyl adenosine; nm5U=5-aminomethyl uridine; A=adenosine and U=uridine. m (subscript) indicates that the RNA strands contained 2′-methoxy nucleotides.
In this experiment, the individual reaction steps were monitored by HPLC (Figure 3b). In the first step, the prebiotic loading of Phe onto the RNA strand ON7, under the reaction conditions described above with GdmCl, gave the donor RNA strand ON8 in ca. 38 % yield. In the second step, we combined the isolated donor RNA strand ON8 with an equimolar amount of the acceptor RNA strand ON9 in 100 mM 2-(N-morpholino)ethanesulfonate (MES) buffer pH 6 and 1 M NaCl. The RNA strand ON9 contained 2′-methoxy nucleotides, which are found in contemporary ribosomal RNA (rRNA)69 and minimize degradation. The addition of 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium chloride (DMTMM⋅Cl) to the solution mixture of ON8 and ON9 at 15 °C led to the formation of the hairpin intermediate ON10 in ca. 40 % yield. Since the melting temperature of the reactive duplex ON8⋅ON9 was ca. 35 °C in the buffered aqueous solution (Figure S20a–b), it was assembled almost quantitatively in the reaction mixture. As reported previously, the activation of the carboxylic acid could also be performed prebiotically with methylisonitrile (MeNC) and 4,5-dicyanoimidazole (DCI).11, 70 In the third step, we warmed the aqueous solution of the hairpin intermediate ON10 to 90 °C at pH 6 to cleave the urea bond,71, 72 which furnished the RNA-peptide chimera ON11 in ca. 48 % yield. In contrast to our previous experiments with donor RNA strands containing amino acid modified N6-methyl-N6-carbamoyl adenosine at the 5′-end,11 the absence of the N6-methyl substituent in ON8 prevented the formation of a hydantoin side-product in the urea cleavage step.73
Finally, we performed the loading of Phe and the RNA-based amino acid transfer in a one-pot experiment (Figure 3c). The crude reaction mixtures were filtered after each step to remove the excess of salts and low-molecular weight compounds but retaining the RNA strands. Indeed, analysis of the one-pot experiment by HPLC showed the formation of the RNA-peptide chimera ON11 in ca. 11 % overall yield (Figure 3c), which was in line with the result obtained for the three individual reaction steps (ca. 7 %).
Conclusion
Recently, we reported that RNA with the help of two non-canonical nucleosides, that is, amino acid modified N6-carbamoyl adenosine and 5-methylaminomethyl uridine, gained the ability to carry amino acids at certain nucleobase positions. The subsequent transfer of amino acids from one nucleobase to another enabled the synthesis of small peptides on RNA, affording RNA-peptide chimeras. The ability of RNA to decorate itself with the help of non-canonical nucleosides constitutes an additional function, which places RNA in a prime position for enabling the chemical evolution of life.
In this work, we described a new method to load RNA strands with amino acids. While this loading reaction was so far only possible with adenosine in low yields, we could now improve the loading efficiencies to yields up to 84 %. Most importantly, the loading reaction was also possible in RNA strands with acceptable efficiencies between 10–23 %. These results suggested that RNA-peptide chimeras are molecules that could have formed under early Earth conditions. In addition, we expanded the loading reaction to N-methylcarbamoyl guanosine and cytidine, and to their corresponding RNA strands. A caveat of the reported process is that we detected deamination of guanosine under the loading conditions. In contrast, the loading with Phe and the subsequent RNA-based amino acid transfer was found to be surprisingly efficient. This observation suggests that loading preferences, for yet unknown reasons, might exist. Thus, we believe that different amino acids might have distinct preferences to be loaded onto specific sequences. This is the prerequisite for RNA-encoded peptide synthesis. Our study also showed that the absence of a N6-methyl substituent in N6-methylcarbamoyl adenosine enhanced the loading efficiency and prevented the formation of hydantoin side-products.
The reported chemistry also required big changes in the pH of the aqueous solution. While the nitrosation of the N6-methylcarbamoyl nucleoside took place under acidic conditions (5 % H3PO4), the reaction with the amino acid was performed at the optimal pH 9.5. Therefore, we need to assume that the first reaction step occurred in an acidic environment, such as an acidic pond, from which a mixture of the nitrosated nucleoside and the amino acids flow out, e.g. along a carbonate containing river bed or into a carbonate dominated second basin. Repetitive loading and RNA-based peptide synthesis cycles would require continuous fluctuations of the pH between acidic and moderately basic in a dynamic process. However, it is worth mentioning that a low amino acid loading efficiency is not necessarily a disadvantage for an RNA-peptide world, since a high turnover of canonical nucleotides into N-carbamoyl derivatives would lead to the loss of the information-encoding property of RNA, hindering replication. We are just at the beginning of learning about the possibilities that are offered by RNA-peptide conjugates and how they could be integrated into coding schemes, e.g. via Hoogsteen-type base-pairing.
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft for supporting this research through the DFG grants: CA275/11-3 (ID: 326039064), CRC1309 (ID: 325871075, A4), CRC1032 (ID: 201269156, A5) and CRC1361 (ID: 393547839, P2). We thank the Volkswagen Foundation for funding this research (grant EvoRib). This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program under grant agreement No. 741912 (EPiR). L. E. thanks the Alexander von Humboldt Foundation for a postdoctoral fellowship (ESP 1214218 HFST-P). We also thank M.Sc. Johann de Graaff for helpful discussions. Open Access funding enabled and organized by Projekt DEAL.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information of this article.