Rapid Formation of Non-canonical Phospholipid Membranes by Chemoselective Amide-Forming Ligations with Hydroxylamines**
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.26434/chemrxiv-2023-02t0k).
Abstract
There has been increasing interest in methods to generate synthetic lipid membranes as key constituents of artificial cells or to develop new tools for remodeling membranes in living cells. However, the biosynthesis of phospholipids involves elaborate enzymatic pathways that are challenging to reconstitute in vitro. An alternative approach is to use chemical reactions to non-enzymatically generate natural or non-canonical phospholipids de novo. Previous reports have shown that synthetic lipid membranes can be formed in situ using various ligation chemistries, but these methods lack biocompatibility and/or suffer from slow kinetics at physiological pH. Thus, it would be valuable to develop chemoselective strategies for synthesizing phospholipids from water-soluble precursors that are compatible with synthetic or living cells Here, we demonstrate that amide-forming ligations between lipid precursors bearing hydroxylamines and α-ketoacids (KAs) or potassium acyltrifluoroborates (KATs) can be used to prepare non-canonical phospholipids at physiological pH conditions. The generated amide-linked phospholipids spontaneously self-assemble into cell-like micron-sized vesicles similar to natural phospholipid membranes. We show that lipid synthesis using KAT ligation proceeds extremely rapidly, and the high selectivity and biocompatibility of the approach facilitates the in situ synthesis of phospholipids and associated membranes in living cells.
Introduction
Phospholipids are amphiphilic species, consisting of a polar headgroup and two fatty acyl chains, that self-assemble into stable bilayers. Phospholipids make up most of living cell membranes, and their central role in life and unique self-assembly properties have led to a multitude of uses for synthetic lipid membranes. For example, researchers have used lipid membranes as drug-delivery carriers, for reconstituting membrane associated proteins, as models for understanding the origin-of-life, and as compartments for synthetic cell models.1-4 While the capability of phospholipids to self-assemble into membranes is well characterized, the de novo synthesis and formation of phospholipid membranes remains both challenging and limited in scope.5 The biosynthesis of phospholipids is mediated by complex enzymatic machinery,6-8 and these enzymes are often difficult to fully reconstitute in vitro, especially without a pre-existing membrane-bound environment.4 An alternative and potentially simpler approach is to develop highly efficient, non-enzymatic and chemoselective strategies for synthesizing phospholipids from water-soluble precursors.9 Previous work has demonstrated that various bioconjugation chemistries can be used to generate phospholipid species in water, but past studies have suffered from one or more of several drawbacks such as lack of biocompatibility of the reactants, the formation of unnatural linkages, or poor reaction kinetics in physiological pH conditions.10-12 These issues have precluded the use of such chemistries to generate synthetic phospholipids in living cells.
In surveying possible bioconjugation chemistries to employ for lipid synthesis, we became interested in seminal studies from the Bode group, that reported on two novel amide bond-forming methodologies, KAHA and KAT ligations, based on the reaction between O-substituted hydroxylamines (HA) and α-ketoacids (KAs) or potassium acyltrifluoroborates (KATs), respectively.13, 14 Both ligations are chemoselective and have been extensively utilized in peptide synthesis and other bioconjugation applications.15-18 The reactions are highly efficient in aqueous conditions19 and do not require additional reagents or catalysts. The KAHA ligation forms amides by decarboxylative condensation of N-alkylhydroxylamines and α-ketocarboxylic acids, producing only water and carbon dioxide as byproducts.20, 21 Alternatively, KAT ligations take advantage of the reaction between hydroxylamines and acyltrifluoroborates to rapidly form amides in high yield. While KAHA and KAT ligations have been shown to occur between a wide range of hydroxylamine substrates and acyl donors, to our knowledge, the application of these bioconjugation reactions to the formation of phospholipid analogs has not been previously studied.
Here, we adapt KAHA and KAT ligations to the synthesis of non-canonical phospholipids in aqueous solution at physiological pH (Figure 1). Both amide bond-forming strategies enable the rapid generation of artificial amidophospholipid 3 in water by the reaction between a hydroxylamine-modified lysophospholipid 1 and an appropriate fatty acyl donor: 2-oxoheptadecanoic acid (2) in the case of the KAHA ligation, and potassium palmitoylacyltrifluoroborate (4) in the case of the KAT ligation (Figure 1). These enzyme-free reactions lead to de novo membrane formation from biocompatible water-soluble precursors, generating cell-like compartments. We characterized the lipid membranes formed using fluorescence microscopy, X-ray diffraction, and cryogenic electron microscopy (cryo-EM). Finally, to demonstrate the bioorthogonality and biocompatibility of both amide-forming lipid synthesis strategies, we conducted the ligations in the presence of cultured mammalian cells and successfully detected the corresponding phospholipid product and imaged the formation of new membranes. Overall, our work demonstrates that KAHA and KAT ligations are highly rapid and selective methods to generate novel lipid membranes, both in vitro and in living cells.
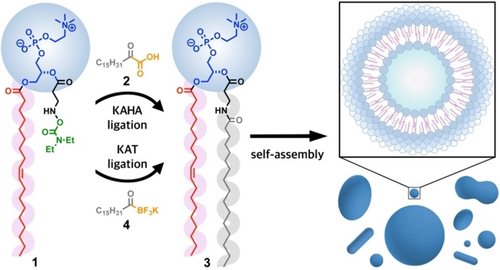
De novo formation of phospholipid membranes using chemoselective amide-forming ligations. Hydroxylamine-modified lysophosphatidylcholine 1 reacts with 2-oxoheptadecanoic acid 2 (KAHA ligation) or potassium palmitoylacyltrifluoroborate 4 (KAT ligation) to generate the non-canonical phospholipid 3, which spontaneously self-assembles into membrane-bound vesicles in water (HEPES buffer, pH 7.3).
Results and Discussion
We initially synthesized the hydroxylamine-containing lysophospholipid derivative 1. Previous studies have shown that the formation of stable nitrones prevents amide-forming reactions between acyltrifluoroborates and O-unsubstituted hydroxylamines.14 Taking this into account, several groups have recently explored a variety of O-substituted hydroxylamine reagents, including O-benzoyl hydroxylamines14 and O-carbamoylhydroxylamines,16, 17 which have been shown to participate in KAT ligations in aqueous media with high reaction rates. Therefore, we initially prepared an O-carbamoylhydroxylamine derivative (Scheme S1), which was incorporated into 1-oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (Lyso C18 : 1 PC-OH) to form 1 (Scheme S2). We chose to modify a lysophospholipid containing an unsaturated chain in the sn-1 position to enhance the fluidity of the membranes that would form after phospholipid formation and self-assembly. For the KAHA and KAT reactive partners (ketoacid 2 and acyltrifluoroborate 4, respectively), we derivatized palmitic acid following standard protocols.22-24 Our expectation was that lysolipid derivative 1 would not form membranes as it only contains one long-chain alkyl tail. After amide-forming ligation with the fatty-acyl KAHA or KAT reagent, a second long-chain tail would be appended resulting in a non-canonical phospholipid analog that would be able to spontaneously self-assemble to form phospholipid membranes.
KAHA ligation was initiated by mixing a solution of 1 (1 mM) with 2 (1 mM) in 2 mM HEPES buffer, pH 7.3 at 37 °C. Phospholipid formation was monitored over time by high-performance liquid chromatography/mass spectrometry (HPLC/MS) combined with evaporative light-scattering detection (ELSD) (Figure 2A,B). Using calibration curves of purified phospholipid 3, we determined that 0.80 mM of phospholipid 3 was formed after 6.5 h, corresponding to a yield of 80 %. We subsequently tested KAT ligation by mixing a solution of 1 (1 mM) with 4 (1 mM) in 2 mM HEPES buffer, pH 7.3 at room temperature (Figure S1). Remarkably, we estimated by HPLC/MS that 0.97 mM of phospholipid 3 was formed at room temperature after 2 min of reaction. Notably, we observed that both ligations proceeded smoothly at different buffer concentrations (Figure S26). To further test the speed of the KAT ligation, we lowered the concentration of reactive precursors to 25 μM and performed the reaction at room temperature. Even under these more stringent conditions, we found that hydroxylamine derivative 1 was fully converted to 3 in the presence of 4 within minutes (Figure 2C,D). The identification of 3 was confirmed by HRMS and comparison to a synthetic standard which showed an identical retention time and mass (Scheme S7, Figure S1). Previous work has established that KAT ligations can proceed much more rapidly than KAHA ligations, however, these reactions normally require mildly acidic conditions.25, 26 The highly rapid rates of reaction at physiological pH, especially for the KAT ligation, might be due to self-assembly of the lipid precursors, which are likely forming mixed micelles that position the reactive hydroxylamines and acylating agents in very close proximity to one another. Previous work has shown that the effective molarity of reagents in mixed micelles can be extremely high, dramatically increasing the rates of otherwise sluggish reactions.9, 27, 28 The critical micelle concentrations (CMC) of lipid precursors 1, 2, and 4 were estimated by a previously reported method29 based on the solvatochromic fluorescent dye Nile Red (Figure S16).
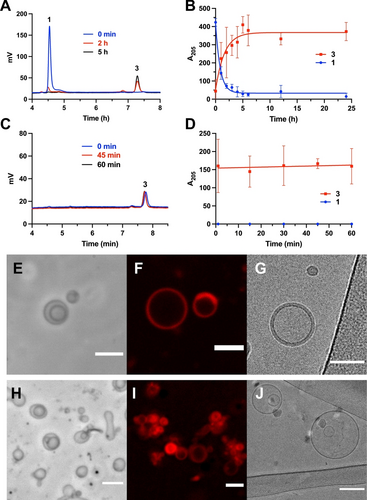
Phospholipid membrane formation induced by KAHA and KAT ligations between the hydroxylamine-modified lysophosphatidylcholine 1 and a palmitoyl donor, 2-oxoheptadecanoic acid 2 for KAHA; potassium palmitoylacyltrifluoroborate 4 for KAT. A) HPLC/ELSD traces monitoring the formation of phospholipid 3 by KAHA ligation at 37 °C between the hydroxylamine derivative 1 (1 mM) and the ketoacid 2 (1 mM). The retention times for all species were verified by mass spectrometry and the use of fully characterized standards. Data was collected in triplicate. B) Reaction between 1 and 2. Kinetic curve of consumption of 1 and production of 3 over 24 h. Error bars represent standard deviations (SD) (n=3). C) HPLC/ELSD traces monitoring the formation of phospholipid 3 by KAT ligation at room temperature between 1 (25 μM) and 4 (25 μM). The retention times for all species were verified by mass spectrometry and the use of fully characterized standards. Data was collected in triplicate. D) Reaction between 1 and 4. Kinetic curve of consumption of 1 and production of 3 over 60 min. Error bars represent standard deviations (SD) (n=3). E) Phase-contrast microscopy image of the spontaneous assembly of phospholipid 3 into vesicular structures after 6 h of KAHA reaction. Scale bar denotes 5 μm. F) Fluorescence microscopy image of vesicles from KAHA reaction mixture stained with 0.2 % mol Nile Red. Scale bar denotes 5 μm. G) Cryo-EM image of vesicles from KAHA reaction mixture. Scale bar denotes 50 nm. H) Phase-contrast microscopy image of in situ formed phospholipid 3 vesicles after 30 min of KAT reaction, Scale bar denotes 5 μm. I) Fluorescence microscopy image of vesicles from KAT reaction mixture stained with 0.2 % mol Nile Red. Scale bar denotes 5 μm. J) Cryo-EM image of vesicles from KAT reaction mixture. Scale bar denotes 100 nm.
Because phospholipid 3 resembles a canonical phosphatidylcholine structurally and chemically, we expected that spontaneous membrane assembly would likely occur leading to de novo bilayer vesicle formation during synthesis. Therefore, we decided to observe the in situ reaction mixture of both KAHA ligation and KAT ligation in 2 mM HEPES buffer, pH 7.3, using microscopy. As expected, we did not observe membranes in the presence of 1, 2, or 4 alone. However, using phase-contrast microscopy, we were able to detect the spontaneous assembly of phospholipid 3 into vesicular structures when the KAHA ligation was performed between 1 and 2 after 6 h at 37 °C (Figure 2E). The diameter of the observed vesicles ranged from 2–10 μm. The lipid vesicular structures were further identified by fluorescence confocal microscopy after adding 0.2 mol % membrane-staining dye Nile Red (Figure 2F). The bilayer nature of the membrane-bound vesicles could be readily observed by cryo-EM (Figure 2G). Similar observations were noted after KAT ligation between 1 and 4 within 30 min, with the difference that more abundant vesicles were observed (Figure 2H–J). To further characterize the vesicle size distribution upon in situ formation of phospholipid 3, we carried out dynamic light scattering (DLS) analyses before and after the two ligations (Figure S14). Notably, the DLS results of dispersions of lysolipid 1 alone indicated the presence of small aggregates with size around 10 nm, likely corresponding to micelles. However, when compound 2 or 4 was added, the DLS profile changed dramatically, showing high polydispersity due to the presence of vesicles ranging from nanometer to micrometer sizes. Such polydispersity can be decreased by filtering the sample through a 0.45 PTFE filter to remove bigger vesicles, which results in a more uniformly dispersed vesicle population.
To further characterize membranes of 3, we hydrated a thin film of purified phospholipid 3 in HEPES buffer, pH 7.3 at room temperature for 30 min, which resulted in the formation of micrometer-sized membrane-bound vesicles. Abundant vesicles were observed after tumbling the mixture at 37 °C (Figure S15). Membrane-bound vesicles were initially identified by fluorescence microscopy using the 0.2 mol % membrane-staining dye Nile Red (Figure 3A). Transmission electron microscopy (TEM) also corroborated the formation of vesicles (Figure 3B). To further characterize the formed structures, we performed cryo-EM (Figure 3C) and could observe the expected lamellar structure of the lipid bilayers. Additionally, we demonstrated that liposomes formed from 3 were capable of encapsulating and retaining polar molecules, such as 8-hydroxypyrene-1,3,6-trisulfonic acid (HPTS) (Figure 3D).
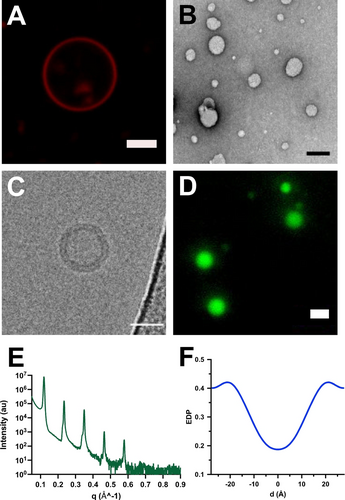
Characterization of vesicles formed from phospholipid 3. A) Fluorescence microscopy image of vesicles formed by hydration of a thin film of 3. Membranes were stained with 0.2 mol % Nile Red. Scale bar denotes 5 μm. B) TEM image of negatively stained 3 vesicles. Scale bar denotes 100 nm. C) Cryo-EM microscopy image of membrane-bound vesicles resulting from the self-assembly of phospholipid 3. Scale bar denotes 50 nm. D) Fluorescence microscopy image demonstrating the encapsulation of HPTS in phospholipid 3 vesicles. Scale bar denotes 5 μm. E) X-ray diffraction (XRD) intensity profile of a multilayer film of 3 at 25 °C with 97 % relative humidity. F) Relative electron density profile (EDP) of a multilayer film of 3 at 97 % relative humidity and 25 °C. 0 Å represents the midplane of the lipid bilayer.
We also conducted X-ray diffraction experiments to better quantify the structure of the bilayers formed by self-assembly of 3. The diffraction measurements were carried out using an in-house Cu Kα tube spectrometer with a wavelength of 1.54 Å operating in the horizontal plane. Multistacks of oriented lipid bilayers were deposited on silicon wafers and the relative humidity (RH) was accurately controlled by a specially constructed humidity cell as previously described.30 X-ray diffraction studies were performed at 98 %, 93.5 %, 83 % and 75 % RH at room temperature (Figure S17). The scattered intensity was plotted as a function of q (the scattering vector), which is directly related to the scattering angle by q=4π sin(θ)/λ, where λ is the wavelength of the X-rays and θ is the incident angle. The diffraction measurements of the assemblies of 3 on the silicon wafer showed 5 equidistant Bragg peaks (Figure 3E), which corresponds to a lipid lamellar phase. The characteristic distance between the lamellar Bragg peaks on the one-dimensional I(q) profile is related to the membrane repeat distance by D=2π/Δq. The lamellar repeat distance (D) denotes the thickness of the lipid bilayer and its surrounding water layer. For lipid 3, D=54.8 Å at 98 % relative humidity and room temperature. In addition, the diffraction peaks were fitted by a Gaussian after background subtraction to determine their area under the peak and obtain the electron density profile (EDP) (Figure 3F), where, for convenience, d=0 Å represents the lipid bilayer midplane. The EDP of the lipid bilayer presents two characteristic maxima attributable to the glycerol backbone of the lipids and a minimum of intensity that corresponds to the methyl terminal groups of the hydrophobic chains. Analysis of the EDP gave an estimate of the lipid bilayer thickness (Dhh=41.0 Å), and the water layer thickness (Dw=13.8 Å). Membranes formed from 3 have a slightly thicker lipid bilayer when compared to the well-studied monounsaturated phospholipid 1-palmitoyl-2-oleoyl phosphatidylcholine (Dhh=38.7 Å)31 and the unsaturated phospholipid dioleoylphosphatidylcholine (Dhh=36.7 Å) at 30 °C, which is expected given the longer acyl tail at the sn-2 position of 3.32
Having demonstrated that KAHA and KAT ligation can be used to rapidly generate amido-phospholipid 3 under physiological pH conditions, we next explored if the chemoselectivity of the amide-forming ligations could be used to generate phospholipids in living cells. Tools that enable the in cellulo generation of specific lipid species have shown application in studying lipid signaling and the roles of specific lipids in organelle function and membrane fusion/fission.33-35 In principle, KAHA and KAT ligations could be used as a bioorthogonal ligation to form amides in living cells. However, previous studies have been hindered by the requirement of lower pH and sluggish kinetics, though recent innovations have led to reagents that can react much faster and in reaction conditions closer to physiological pH conditions.19 Since we observed such rapid kinetics using lipid modified reactive precursors, even with submillimolar concentrations and at physiological pH, we speculated that lipid formation could be compatible with live cells, enabling the generation of phospholipid analogs in cell membranes. To trigger the in situ synthesis of phospholipid 3 by KAHA ligation in HeLa cells, solutions of hydroxylamine derivative 1 and the ketoacid 2 in cell media (DMEM) were added to plates containing confluent HeLa cells. Cells were first incubated with 1.5 mL of 500 μM 1 in DMEM for 1 h at 37 °C, followed by addition of 1.5 mL of 500 μM 2 in DMEM. The final concentration of each lipid was 250 μM. After 6 h of incubation at 37 °C, the media was removed, and the cells were washed with Hank's balanced salt solution (HBSS) and the cellular lipids were extracted using a modified Folch method.36 We next redissolved the lipid extract to a concentration of 1 mg/mL in MeOH/CH2Cl2 (95 : 5) and analyzed it by HPLC/MS running in single ion monitoring (SIM) mode (Figure 4A). The retention time for in situ formed phospholipid 3 was verified by mass spectrometry ([MH]2+=416.3) and the use of previously synthesized phospholipid 3 as a known standard. In addition, we also tested a sequential incubation where we first exposed HeLa cells to ketoacid 2 for 1 h at a concentration of 500 μM. After cell medium removal, DMEM containing 500 μM of 1 was added to the cell, followed by continued incubation for an additional 6 h. HPLC/MS experiments detected the product 3 from the cellular lipid extracts. We next tested if the KAT ligation could proceed in live cells. Considering the higher reactivity and efficiency obtained from the in vitro KAT ligation, we decided to decrease the concentration of the reactive precursors (1 and 4) and the incubation time for the cellular KAT ligation. HeLa cells were incubated in DMEM containing 200 μM 1 for 1 h at 37 °C followed by addition of 200 μM 4 in DMEM to reach a final concentration of 100 μM for both 1 and 4. Cells were incubated with both lipids for 2 h at 37 °C. After cell lysis and lipid extraction, detection of the phospholipid 3 mass peak was successfully observed (Figure 4A). To discard the possibility that the observed compound 3 was formed in the extracellular environment and subsequently entered cells, we conducted experiments in which we incubated HeLa cells with DMEM containing 500 μM of purified 3 for 6 h at 37 °C. HPLC/MS analysis of the lipid extract obtained after cell media removal and cell lysis showed the absence of phospholipid 3 in the extract (Figure S18). This absence might be attributed to the tendency of phospholipid 3 to form vesicles, external to the cells, which do not fuse with the cell membrane. Conversely, our results suggest that single chain amphiphile precursors can readily integrate into the cell membrane. Finally, control experiments in which we omitted addition of reactive lipid precursors led to no detectable 3 in HeLa cell lipid extracts, which is expected given that 3 is a non-canonical phospholipid. While it is possible that the lipid precursors could be consumed over time by cellular metabolism or degradation, our data indicate that such consumption does not prevent the formation of phospholipid 3. Our results demonstrate that both KAHA and KAT amide-forming ligations can occur in living cells within hours of incubation with lipid precursors.
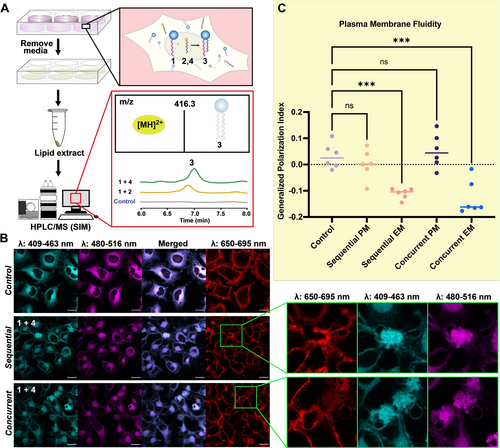
Live-cell experiments with KAHA and KAT ligations A) Flowchart of in vivo experiment using HeLa cells. Lipids were extracted using a modification of Folch's method and analyzed by HPLC/MS employing single ion monitoring (SIM) mode. Phospholipid 3 formation was detected [MS(ESI): 416.3 ([MH]2+)] in cellular reactions of KAHA and KAT ligations, while no signal was detected in the control experiment when lipid precursors were not added to HeLa cells. B) Confocal microscopy images of cells stained with 0.1 vol % of stock solution (2 mg/mL) of C–Laurdan dye and 0.1 vol % of stock solution (2 mg/mL) CellMaskTM plasma membrane stains. Images were collected between 409 to 463 nm (cyan) for the lipid ordered channel, 480–516 nm (magenta) for the lipid disordered channel, and 650–695 nm for the plasma membrane stain (red). Merged images of the ordered and disordered channels are also shown. Magnified inset highlights excess membrane structures observed in the sequential and concurrent experiments. C) C–Laurdan dye generalized polarization (GP) indexes at each pixel position in the plasma membrane (PM) region and excess membrane (EM) region in HeLa cells with 1 and 4 added sequentially and concurrently. (N=6), *** indicates significant differences from the control treatment with p 0.001. ns indicates no significant difference.
To further understand the biocompatibility of the ligation reactions, we performed live-cell imaging using confocal microscopy to observe the morphological changes that occurred in the cell membrane upon addition of lipid precursors. Cells were imaged in three conditions (Figure 4B): HeLa cells without lipid precursors added as a control; HeLa cells incubated with medium containing 100 μM 1 for 1 h, which was removed before addition of medium containing 100 μM 4, then incubated for 2 h with 4 (labeled as sequential); HeLa cells pre-incubated with 200 μM 1 for 1 h, followed by addition of 200 μM 4 to reach a final concentration of 100 μM of each lipid, followed by incubation for 2 h (labeled as concurrent). CellMaskTM staining was also performed to confirm high plasma membrane (PM) integrity (Figure 4B). The lack of membrane disruption further demonstrated the biocompatibility of 1 and 4. Cells were also stained with C–Laurdan, a solvatochromic fluorescent membrane dye. C–Laurdan exhibits spectral sensitivity to the polarity of its binding region, resulting in its emission spectra in a polar environment to be red-shifted by approximately 50 nm. Loosely packed membranes allow for a higher penetration of water molecules, which leads to higher effective polarity,37 and for this reason the emission spectra of C–Laurdan in disordered membranes is red-shifted with respect to more ordered membranes. C–Laurdan allows the visualization of ordered lipid membranes between the wavelengths of 409 to 463 nm (cyan), whereas more disordered membranes are observed between wavelengths of 480–516 nm (magenta). The images from the different wavelengths were merged (Figure 4B, merged) to provide a visual representation of the relative ratio of ordered and disordered membranes. The shift in emission profile can be quantified by the generalized polarization (GP) index, which is a ratiometric measurement of the fluorescence intensity in the ordered and disordered channel. We calculated the generalized polarization indexes of the plasma membrane based on a previously described method38 from the sequential and concurrent experiments and found it showed no significant difference from that of the control, indicating they have comparable levels of lipid order or fluidity (Figure 4C, sequential and concurrent PM). This data suggests that the reaction between 1 and 4 to form 3 results in minimal alteration of the lipid order and fluidity of the plasma membrane in HeLa cells.
Interestingly, we observed numerous vesicular membrane structures budding out of the cell membrane when 1 and 4 are both added to cells, either sequentially or concurrently (Figure 4B, magnified inset). Given that the reaction is occurring in cells (Figure 4A), we speculated that the membrane structural changes are likely being induced by generation of 3. We further hypothesized that the accumulation of 3 leads to excess membrane (EM) formation within the cells, and this manifests itself through the formation of additional membrane-bound structures that are not normally observed. We calculated the GP index of the excess membrane portion and compared it to that of the surrounding plasma membrane area. The GP index of the excess membrane is significantly less than the plasma membrane, indicating higher membrane fluidity (Figure 4C, sequential and concurrent EM). This is likely due to 3 being an unsaturated phosphatidylcholine, which prevents tight packing of the lipids with one another and results in higher fluidity.39 The potential lower abundance or lack of cholesterol, sphingolipids, and membrane proteins in the newly formed membrane of 3 is another explanation for the lower observed GP index.40, 41 Our observation of excess membrane formation suggests that the cell membrane can be remodeled using non-canonical lipids and KAT bioorthogonal ligation chemistries.
Conclusion
In summary, we have demonstrated that KAHA and KAT ligations can be used for the in situ synthesis of membrane-forming non-canonical phospholipids from water-soluble precursors. When applied to the ligation of lipid fragments, these amide-forming ligations are highly efficient and proceed rapidly in physiological pH conditions. This work expands the toolbox of reactions that trigger de novo phospholipid vesicle formation,9, 10, 42-44 and paves the way for advanced synthetic cell development and possible multiplexing of mutually orthogonal bioconjugation chemistries to generate more complex multicomponent synthetic membranes.5, 45, 46 While KAHA ligation using ketoacids is probably not chemoselective in live cells due to the expected quantities of competing ketoacids, we found that lipid conjugation could indeed proceed within cell membranes, highlighting its potential for artificial lipid synthesis. Notably, we observed a significant enhancement in amide formation during KAHA and KAT ligations conducted under physiological pH conditions. These findings are most likely a consequence of the hydrophobic interactions among the various molecular components involved in the reactions, which underscores the importance of considering the influence of hydrophobic associations in designing and optimizing chemical reactions. When lipid ligations were performed in living cells, membrane morphological and fluidity changes were observed. These results suggest that our approach could potentially be used to engineer new membranes within cells and understand the effect of lipid composition on membrane behavior in living cells,47 which is very difficult to probe genetically. Beyond generating non-canonical lipid species, by further extending the types of hydroxylamine and acylating probes, we envision that KAHA/KAT lipid bioconjugation could be employed to control the formation of naturally occuring amide-containing lipids such as lipoamino acids and sphingolipids.
Acknowledgments
This work was supported by the National Institutes of Health (R35GM141939), the Agencia Estatal de Investigación (AEI) and the Ministerio de Ciencia e Innovación (MICINN) [PID2021-128113NA-I00]. R.J.B. also thanks the Agencia Estatal de Investigación (AEI) and the Ministerio de Ciencia e Innovación (MICINN) for his Ramón y Cajal contract (RYC2020-030065-I). I.B. acknowledges support from the National Science Foundation (MCB-2046303). The authors acknowledge the facilities, along with the scientific and technical assistance of the Dr. Mariusz Matyszewski from the cryo-EM facility and Dr. Yongxuan Su from the Molecular Mass Spectrometry Facility at UC San Diego.
Conflict of interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.