Genetic testing for inherited retinal degenerations: Triumphs and tribulations
Abstract
Inherited retinal degenerations (IRDs) are a genotypically and phenotypically diverse group of conditions. Great strides have been made toward identifying the genetic basis for these conditions over the last 30 years—more than 270 different genes involved in syndromic and nonsyndromic forms of retinal dystrophies have now been identified. The identification of these genes and the improvement of clinical laboratory techniques have led to the identification of the genetic basis of disease in 56–76% of patients with IRDs through next generation sequencing and copy number variant analysis. Genetic testing is an essential part of clinical care for patients affected with IRDs and is required to confirm the diagnosis, understand the inheritance of the condition, and determine eligibility for gene-specific treatments or clinical trials. Despite the success achieved in determining the genetic cause of these conditions, several challenges remain, which must be considered when providing genetic testing and genetic counseling to patients. For this reason, an integrated team of ophthalmic and genetic clinicians who are familiar with these challenges is necessary to provide optimal comprehensive care to these patients.
1 INTRODUCTION
Inherited retinal degenerations (IRDs) are a phenotypically and genotypically diverse group of conditions that cause vision loss due to a loss of function of retinal photoreceptor cells. Presenting symptoms may be poor peripheral and/or night vision, as seen in retinitis pigmentosa (RP); poor central vision, as seen in Stargardt disease; or early onset nystagmus, as seen in Leber congenital amaurosis (LCA), and achromatopsia. These conditions may present as isolated ophthalmic findings or in the context of a syndrome. The most common IRD is RP or rod-cone dystrophy (RCD), with an incidence of 1/3,500 (Haim, 2002). While individually each of these IRDs is rare, collectively it is estimated that there are more than 140,000–200,000 people affected with retinal dystrophies in the United States (Daiger, Sullivan, & Bowne, 2013; Stone et al., 2017). A patient presenting to a genetics clinic may have an existing diagnosis of a macular dystrophy, cone-rod dystrophy, or RCD. These are general categories of IRDs, but also frequently overlapping in phenotype. By ordering genetic testing on these patients through a broad inherited retinal dystrophy panel, a more specific diagnosis, information on prognosis, additional information on inheritance of the condition, and potential eligibility for clinical trials may be obtained.
Part of the complexity of these conditions is that multiple genotype–phenotype associations have been established for them. A single phenotype may arise from a variety of different genetic causes, and, conversely, a single gene may be associated with multiple phenotypes (Berger, Kloeckener-Gruissem, & Neidhardt, 2010). The PRPH2 gene is a prime example of this phenotypic heterogeneity: pathogenic variants in this gene may be associated with macular disease such as pattern dystrophy, butterfly macular dystrophy, or vitelliform dystrophy, or it may cause peripheral retinal degeneration such as RP (Leroy, Kailasanathan, De Laey, Black, & Manson, 2007). This variability may even be seen within the same family. In other cases, pathogenic variants in the same gene may cause some patients to be affected with syndromic disease and others with nonsyndromic disease. The USH2A gene, for example, is the most common cause of nonsyndromic RP and also the most common cause of Usher Syndrome Type II (Pontikos et al., 2020; Stone et al., 2017).
The first IRD-associated gene (RHO) was identified in 1990 (Dryja et al., 1990). At that time, it was anticipated that a small number of genes would be found to cause IRDs. However, more than 270 IRD-related genes have now been identified, including 90 for nonsyndromic RP alone, as shown in Figure 1 (https://sph.uth.edu/retnet/). While disease-causing variants are much more prevalent in some of these genes are than in others, there is no single genetic cause that is responsible for the majority of these patients. A large genotyping study of over 3,000 families affected with IRDs in the United Kingdom identified molecular diagnoses due to pathogenic variants in 135 different genes; however, 70% of families had pathogenic variants in the most common 20 genes (Pontikos et al., 2020). This study and a similar study in the United States found that the three most common genetic causes of IRDs in their study populations were ABCA4, USH2A, and RPGR (Pontikos et al., 2020; Stone et al., 2017).
While genetic testing for IRDs is supported by large organizations such as the American Academy of Ophthalmology as an important component of clinical care, financial, and logistical barriers prevent widespread genetic testing (Erwin, LaMaire, Espana, Eble, & Dhar, 2020; Harrison et al., 2015; Stone et al., 2012). In order to overcome this, several different genetic testing initiatives have been developed by government programs, nonprofit foundations, and corporations in which genetic testing through a CLIA-certified lab is available to patients at no cost to them. These programs include the eyeGENE research study at the National Eye Institute, the Foundation Fighting Blindness' My Retina Tracker Genetic Testing Study and Open Access Genetic Testing Program, and Spark Therapeutics' ID Your IRD Program.
The eyeGENE research study collected more than 6,000 samples from 2006 to 2015 from patients and family members affected with genetic eye diseases, including IRDs (Parrish et al., 2016). Through this research initiative, patients provided DNA samples and phenotypic data to a biorepository (Goetz, Reeves, Tumminia, & Brooks, 2012). Genetic testing was performed on the samples and returned to patients via their ordering providers. The samples and information in the repository were made available to researchers through an application process.
In 2017, Foundation Fighting Blindness, a nonprofit patient advocacy organization for patients affected with IRDs, developed a genetic testing study for patients enrolled in their patient registry: the My Retina Tracker Registry (Shaberman & Durham, 2019). Patients provide demographic, family, and medical information for the registry and get genetic testing at no cost through a CLIA-certified lab. Genetic counseling is provided to all participants as part of the program. The program expanded in October 2019, with the development with an Open Access Genetic Testing Program, available to a greater number of patients across the country (https://www.fightingblindness.org/open-access-genetic-testing-program).
The ID your IRD program (https://www.eyewant2know.com/idyourird), sponsored by Spark Therapeutics, the manufacturer of an FDA-approved gene therapy treatment, serves as a similar genetic testing program. The ID your IRD program provides genetic testing at no cost to the patient through a CLIA-certified lab and genetic counseling is available as well. While these programs have differences in testing panels used and data sharing, collectively, these three programs have achieved a goal of making genetic testing available to thousands of patients affected with IRDs in the United States, for whom genetic testing might otherwise have been cost-prohibitive.
2 CURRENT DETECTION RATES
In the field of IRDs, currently available commercial panels and research investigations have greatly enhanced the success of clinicians in identifying the genetic basis for disease. Large scale panels are currently available in commercial testing labs, which vary by the number of genes tested, methodology used, and diseases covered. Deletions and duplications significantly contribute to disease burden, being responsible for 7–9% of pathogenic variants in IRD patients (Ellingford et al., 2018; Zampaglione et al., 2020). Multiple studies have been conducted on large numbers of families (individual studies ranging in size from 700 families to 2,420 families) affected with IRDs in Ireland, Israel, England, and the United States. These studies show an overall detection rate of 56–76% (Carss et al., 2017; Sharon et al., 2020; Stone et al., 2017; Whelan et al., 2020). Clinical detection rates may vary by diagnosis, as described by Carss et al, where detection rates varied from 29% in patients with cone dystrophy to 89% in patients with LCA (Carss et al., 2017). Similarly, studies have also shown that detection rates may vary based on age—with a higher detection rate in individuals less than 50 years of age. (Shah et al., 2020).
3 DEVELOPMENT OF GENE-BASED TREATMENTS AND TRIALS
The field of retinal genetics has proven to be a pioneer in the area of gene therapy when a retinal degeneration was the first disease for which a gene therapy treatment became FDA-approved. This therapy for RPE65-associated retinal degeneration was developed by Spark Therapeutics. Voretigene neparvovec-rzyl (Luxturna) was approved by the FDA to treat patients affected with retinal degeneration due to pathogenic variants in RPE65 in 2017, and the first patient was treated in 2018 (Ciulla, Hussain, Berrocal, & Nagiel, 2020; Russell et al., 2017). Several other gene-based treatments are currently in clinical trials, including gene augmentation for CNGA3, CNGB3, CHM, and RPGR; CRISPR gene editing for CEP290; and oligonucleotide therapy for CEP290, USH2A, and RHO (Thompson et al., 2020). The development of these gene-dependent treatments highlights the importance of determining the specific genetic cause of disease in patients with IRDs.
4 COMPLICATIONS THAT CAN EMERGE WHEN ORDERING GENETIC TESTING
4.1 Identification of unexpected syndromic conditions in patients with isolated retinal dystrophy
There are several syndromes that cause disease in multiple organ systems in addition to the retina (Werdich, Place, & Pierce, 2014). In some of these conditions, the extraocular features of disease are obvious prior to the onset of retinal findings. Two of the more common syndromic retinal dystrophies are Usher Syndrome and Bardet-Biedl Syndrome (BBS). Patients with Usher Syndrome type 1 and 2 have congenital hearing loss and develop RP as a child or young adult. In patients with BBS, the primary features are obesity, postaxial polydactyly, renal disease, hypogonadism, learning disabilities, and RCD. Retinal degeneration is present in more than 90% of these patients (Forsythe & Beales, 2013). Although the diagnosis of BBS may not be suspected until the retinal findings are diagnosed, one or more of the extraocular features of this condition may be present from birth and/or early childhood.
In other cases, the retinal findings may be the presenting sign, or subclinical findings in other organ systems may be present but unknown to the patient and provider. For example, in the juvenile form of neuronal ceroid lipofuscinosis (Batten disease), the typical presentation is a rapidly progressing retinal dystrophy. The vision loss is followed by cognitive decline and other neurodegenerative symptoms (Adams, Mink, & University of Rochester Batten Center Study, 2013) and is characterized by seizures. In cases such as these, as highlighted in the case example below, genetic testing may identify a syndromic retinal dystrophy in a case otherwise suspected to be nonsyndromic.
4.1.1 Case example 1
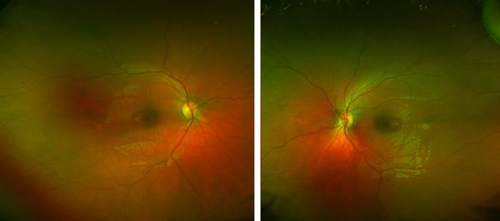
A 6-year-old boy presented for an evaluation for macular dystrophy. No developmental or health concerns were noted, and he was doing well in school. Visual acuity was 20/50 in each eye. The electroretinogram (ERG) was abnormal, with rod responses reduced to 10% of normal and cone isolated responses approximately 40% of normal. On fundus exam, he was noted to have macular atrophy in both eyes (Figure 2). Genetic testing on a retinal dystrophy panel was performed, and he was suspected to have a deletion in the CLN3 gene. Further CNV analysis confirmed that he was homozygous for a 1-kb deletion of the deletions 8–9 (estimated breakpoints chr16: 28498251–28497286; hg19) in the CLN3 gene. This deletion is the most commonly identified pathogenic variant in individuals with CLN3-related juvenile-onset Batten disease. Following the diagnosis, the patient was referred to a multidisciplinary clinic for Batten disease, and he continued to follow in our clinic as well. At the age of 9, his visual acuity had progressed to hand motion vision in the right eye and 20/2,800 in the left eye, which is legally blind, and he was receiving therapy for behavioral problems. Surprising and devastating diagnoses such as this highlight the importance of pretest counseling for patients undergoing large retinal dystrophy panels and specifically of warning them that, even if their condition appears to be solely vision-related, syndromic conditions are also tested on the panel and may be identified.
4.2 Challenges of inheritance counseling
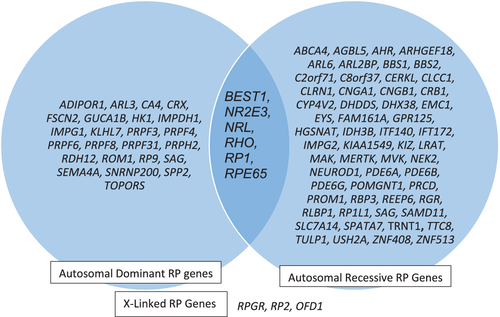
The extreme genetic heterogeneity, multiple inheritance patterns, and diverse phenotypic features of several IRDs can pose challenges for providing accurate genetic counseling and recurrence risks prior to obtaining genetic testing. In the case of nonsyndromic RP, over 90 genes have been identified, which have autosomal dominant, autosomal recessive, X-linked, and mitochondrial forms of inheritance (https://sph.uth.edu/retnet/). Some of these RP genes, including BEST1, NR2E3, NRL, RHO, RP1, and RPE65 (Daiger, Bowne, & Sullivan, 2014) may be associated with both autosomal dominant and autosomal recessive patterns of inheritance (Figure 1). Several of the retinal dystrophy genes may be associated not only with variable inheritance patterns, but also with variable phenotypes. As an example of this, the CRX gene may be associated with autosomal dominant cone-rod dystrophy, autosomal dominant or autosomal recessive LCA, and autosomal dominant RP (Sohocki et al., 1998). There has also been a report of a family with a child affected with LCA with a homozygous pathogenic variant in CRX, and her carrier parents manifested milder late onset retinal abnormalities (Swaroop et al., 1999).
Pedigree analysis for determining the pattern of inheritance within a family affected with RP is complicated by several factors. In cases of simplex RP, in which no other cases of RP are identified in the family, autosomal recessive inheritance is the most likely pattern of inheritance; however, several studies have demonstrated that these cases may actually be due to mutations in genes with X-linked or autosomal dominant inheritance. In one study, 15% of isolated males affected with RP and cone-rod dystrophies were found to have disease causing variants in the X-linked RPGR gene (Branham et al., 2012). It was not determined if these males harbored de novo mutations or if the variant was passed through unaffected female carriers. Churchill et al. (2013) found that 8.5% of families originally thought to have an autosomal dominant pattern of inheritance were subsequently found to have disease causing variants in the X-linked genes RPGR or RP2. X-linked inheritance should be considered as a possibility in families with more than one generation of individuals affected but where male-to-male transmission is lacking, especially if males are more severely affected than females or if females have asymmetric disease (Churchill et al., 2013). In IRD genes with autosomal dominant inheritance, reduced penetrance can be seen, making pedigree interpretation of these families difficult as well (Rivolta et al., 2006). In particular, pathogenic variants in the splicing factor PRPF31 are associated with nonpenetrance the majority of families (Rose & Bhattacharya, 2016). Furthermore, consanguinity or high carrier rates can create the appearance of dominance in a pedigree when the pattern of inheritance is actually recessive—sometimes termed “pseudo-dominance.” Pseudo-dominant inheritance has been reported in several nonconsanguineous families affected with ABCA4-related retinal dystrophies such as Stargardt disease and cone-rod dystrophy due to the relatively high carrier rate of pathogenic variants in the ABCA4 gene (Huckfeldt, East, Stone, & Sohn, 2016; Maugeri et al., 2000) .
Further complicating inheritance counseling of patients with IRDs is the fact that there may be more than one genetic cause of disease within a family. Typically, it would be assumed that multiple members of a family affected with retinal degeneration would have the same genetic cause of disease. However, there have been reports of more than one genetic cause of retinal degeneration being identified within a family (Birtel et al., 2020; Jones et al., 2017). This situation can complicate inheritance counseling and selection of the appropriate genetic test for individuals in these families.
4.2.1 Case example 2
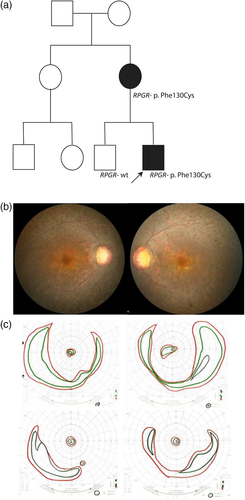
A 9-year-old boy presented to clinic and was diagnosed with RP based on findings of nearly nonrecordable rod and cone ERG responses, elevated dark adaptation thresholds, and peripheral pigmentation in both eyes. At the time of diagnosis, he was believed to be an isolated case of disease. Subsequently, his mother reported night vision loss just before the age of 40. She was found to have significantly reduced rod and cone function. Best corrected visual acuity was 20/25 in her right eye and 20/30 in her left eye when examined at the age of 40. She also had a ring scotoma on visual field testing, and fundus exam showed diffuse retinal atrophy (Figure 3a,b). She subsequently experienced a progressive loss of visual acuity and visual field (Figure 3b). At the age of 45, her visual acuity had progressed to 20/50 in the right eye and 20/100 in the left. Based on the pedigree (Figure 3c), an autosomal dominant pattern of inheritance might have been expected, but the mother and son were found to have a pathogenic missense variant (p.Phe130Cys) in the X-linked RPGR gene, which is believed to be the cause of disease in the family.
4.3 Importance of phenotyping
Since inherited retinal dystrophies are genotypically and phenotypically diverse, with symptoms overlapping between several different conditions, detailed phenotypic information can be important to appropriately guide genetic testing. For example, congenital nystagmus can be seen in several different types of retinal dystrophy, which can have very different prognoses. Retinal dystrophies included in the differential diagnosis for a patient with congenital nystagmus could be LCA, achromatopsia, and congenital stationary night blindness (Papageorgiou, McLean, & Gottlob, 2014). However, since these conditions represent distinct disease categories, they may not be tested on the same genetic panel. Further clinical evaluation, such as an ERG, which may only be available in retinal dystrophy clinics, may be warranted to help clarify the clinical diagnosis. This information is essential in the determination of which genetic test is most appropriate for such patients and in the interpretation of genetic test results. In many cases, testing for a broader retinal dystrophy may assist the geneticist in navigating the diverse phenotypic landscape of these diseases.
4.3.1 Case example 3
A 10-month-old boy was referred to the IRD clinic for a possible diagnosis of LCA based on his history of nystagmus and decreased vision. He was born from a triplet pregnancy, with no history of vision loss in his siblings. His nystagmus was noted at 2 months of age, and his parents reported that he did not follow large objects. In clinic, ERG testing demonstrated near normal rod functioning and near nonrecordable cone functioning. Therefore, the clinical exam was actually consistent with a cone dystrophy such as achromatopsia. This was confirmed with genetic testing, which identified two pathogenic variants in the CNGB3 gene, consistent with this diagnosis. If genetic testing had been ordered for a targeted LCA panel prior without clinical testing to fully evaluate the cause of his vision, this gene would not have been evaluated, and the clinical and genetic diagnosis would have remained unknown for this patient.
5 CONCLUSION
Since the identification of the first retinal dystrophy gene in 1990, significant advances have been made in the study of the genetic basis for inherited retinal diseases. Advances in genetic diagnostic testing have allowed the field to progress to the point where the genetic basis of disease is identified for 56–76% of patients when tested on large NGS panels. The importance of genetic testing is highlighted by the genotypic complexity of this group of conditions, which can only be clarified through testing. This complexity is evidenced not only by the sheer number of genes that have been identified, but also by the wide phenotypic variability and the significant genetic heterogeneity for many of the conditions, which can result in the misinterpretation of inheritance pattern when predicted based only on family history information. In addition, with the development of therapeutic FDA-approved treatments and gene therapy clinical trials, genetic testing results have become essential for the treatment and management of these eye conditions.
Genetic testing has traditionally been performed in the clinical genetics setting, but as it becomes more common in different medical specialties, such as ophthalmology, appropriate integration of this testing becomes necessary. Some IRD clinics have both ophthalmologists trained in IRD and genetic counselors and/or clinical geneticists who work together as a team. However, many ophthalmologists do not have genetic resources available. While ophthalmologists have extensive training in the diagnosis and ophthalmic management of these conditions, they may be less experienced with the nuances associated with ordering genetic testing, interpreting these genetic test results, and genetic counseling of patients affected with inherited retinal conditions. On the other hand, clinical geneticists and genetic counselors would not be able to perform extensive ophthalmic phenotyping in their genetics clinics. As evidenced by the case examples here, there are challenges that exist in selecting the appropriate test for patients, understanding the inheritance of inherited retinal dystrophies, and preparing for unexpected genetic testing results with which genetic and ophthalmic providers must be familiar in order to provide appropriate comprehensive clinical care for these patients.
ACKNOWLEDGMENTS
The authors wish to acknowledge the patients they strive to help, from whom they continuously learn more about their field.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or the article describes entirely theoretical research.