OPTN p.Met468Arg and ATXN2 intermediate length polyQ extension in families with C9orf72 mediated amyotrophic lateral sclerosis and frontotemporal dementia
Abstract
We have ascertained two families affected with familial amyotrophic lateral sclerosis (ALS) in which they both carry a hexanucleotide repeat expansion in the C9orf72 gene, specifically in individuals who also presented with frontotemporal dementia (FTD) or behavioral variant FTD (bvFTD). While some reports attribute this phenotypic heterogeneity to the C9orf72 expansion alone, we screened for additional genetic variation in known ALS-FTD genes that may also contribute to or modify the phenotypes. We performed genetic testing consisting of C9orf72 hexanucleotide expansion, ATXN2 polyglutamine (polyQ) expansion, and targeted next generation sequencing using the ONDRISeq, a gene panel consisting of 80 genes known to be associated with neurodegenerative diseases such as ALS, FTD, Alzheimer's disease, Parkinson's disease, and vascular cognitive impairment. In addition to the C9orf72 expansion, we observed an ATXN2 polyQ intermediate length expansion, and OPTN p.Met468Arg in patients who exhibited ALS and FTD or bvFTD. We conclude that the C9orf72 expansion likely explains much of the ALS-FTD phenotype; however, inheritance of these additional variants likely modifies the disease course and may provide further evidence for biologically relevant oligogenic inheritance in ALS.
1 INTRODUCTION
Based on clinical and genetic studies, it has been proposed that amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are part of the same neurodegenerative disease continuum (Hardiman, van den Berg, & Kiernan, 2011; Woolley & Strong, 2015). ALS is a progressive adult-onset neurodegenerative disorder in which the loss of both upper and lower motor neurons leads to a relentless weakening of limb, bulbar, and respiratory muscles. Death usually ensues within 3–5 years of onset (Renton, Chio, & Traynor, 2014). In contrast, FTD is characterized by progressive neuropsychological deficits, including cognitive, and behavioral dysfunction (Strong et al., 2017). Survival is generally on the order of 5–10 years (Rohrer et al., 2015). There is increasing recognition that ALS and FTD can occur in the same individual, and that both disorders are syndromic with multiple potential etiologies, many of which are genetic in origin (Hardy & Rogaeva, 2014; Ittner et al., 2015). In general, approximately 15% of ALS or FTD cases will progress to develop both phenotypes (Hardy & Rogaeva, 2014; Ittner et al., 2015). This observation underlies the current conceptualization that ALS and FTD are two points on a continuum of a single disorder.
The discovery that a large proportion of ALS and FTD cases harbor a pathogenic hexanucleotide expansion in C9orf72 provides support for this hypothesis (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Rohrer et al., 2015). However, this does not fully explain the significant clinical heterogeneity among patients with similar C9orf72 expansion profiles. Families in which similar C9orf72 expansions are present, but in which there is clinical heterogeneity both within families and between pedigrees, can provide insight into this question. However, until recently the finding of a single genetic variant led to a cessation of the search for additional genetic variants. This reductionist approach has generally constrained the ascertainment of the full extent of genetic mutations in ALS and FTD. However, when such an approach has been used, it has supported the concept of digenic (oligogenic) inheritance (results of study findings summarized in Table 1 and reviewed by Lattante, Ciura, Rouleau, & Kabashi, 2015) (Bury et al., 2015; Cady et al., 2015; Chio et al., 2012; Cooper-Knock et al., 2012; Ferrari et al., 2012; Kaivorinne et al., 2014; King et al., 2013; Lashley et al., 2014; Lattante et al., 2012; Lattante et al., 2015; Lattante, Millecamps et al., 2014; Mignarri et al., 2014; Millecamps et al., 2012; Origone et al., 2015; Pottier et al., 2015; Ratti et al., 2012; Tarlarini et al., 2015; van Blitterswijk et al., 2013; van Blitterswijk, van Es, Hennekam, et al., 2012; van Blitterswijk, van Es, Koppers et al., 2012; van der Zee et al., 2014). Furthermore, there is increasing evidence that additional variation within genes that act on the same pathways as C9orf72 may be modifying the phenotypes of patients (Dekker et al., 2016; Lattante, Le Ber et al., 2014; van Blitterswijk, Mullen, Heckman et al., 2014; van Blitterswijk, Mullen, Wojtas et al., 2014).
| Study | Diagnosis | Other symptoms/alternative diagnosis | Site of onset | Sex | Age at onset (years) | Disease duration (months) | Ethnic origin | C9orf72 expansion | Other variation |
|---|---|---|---|---|---|---|---|---|---|
| Chio et al. (2012) | ALS | FTD | Bulbar | M | 43 | Sardinia | Yes | TARDBP p.A382T | |
| ALS | Mild FTD | Spinal | F | 69 | 43 | Sardinia | Yes | TARDBP p.A382T | |
| ALS | Flail arm variant | Upper limb | M | 35 | Sardinia | Yes | TARDBP p.A382T | ||
| Cooper-Knock et al. (2012) | ALS | Limb | F | 37 | 58 | Yes | TARDBP p.A321V | ||
| ALS | Bulbar | F | 62 | 24 | Yes | FUS p.G174del& | |||
| ALS | Bulbar | F | 50 | 28 | Yes | OPTN p.E322K | |||
| Ratti et al. (2012) | ALS | 47 | Italian | Yes | TARDBP p.A382T | ||||
| ALS | 70 | Italian | Yes | PRPH p.R133P | |||||
| Millecamps et al. (2012) | ALS | Bulbar | 40 | 14 | French-European | Yes | FUS p.R521C | ||
| ALS | Upper limb | 59 | 42 | French-European | Yes | SOD1 p.D110Y | |||
| ALS | 47 | French-European | Yes | ANG p.K17I | |||||
| ALS | 46 | 21 | French-European | Yes | OPTN p.D128EfsX22 | ||||
| ALS | 52 | French-European | Yes | UBQLN2 p.G502_I504del | |||||
| ALS | 42 | French-European | Yes | DAO p.R38H | |||||
| Ferrari et al. (2012) | FTD | 64 | Yes | GRN p.Y294C | |||||
| FTD | 68 | Yes | PSEN2 p.I146V | ||||||
| Lattante et al. (2012) | ALS | 38 | Yes | ANG p.I46V | |||||
| van Blitterswijk, van Es, Hennekam, et al. (2012) | ALS | NL | Yes | VAPB p.V234I | |||||
| van Blitterswijk, van Es, Koppers et al. (2012) | ALS | Cervical | M | 42 | >91 | NL | Yes | TARDBP p.N352S | |
| ALS | Cervical | F | 47 | >15 | NL | Yes | TARDBP p.N352S | ||
| ALS | Cervical | F | 51 | 77 | NL | Yes | SOD1 p.D90A | ||
| ALS | Lumbosacral | F | 53 | >24 | NL | Yes | (1) FUS p.R521C (2) ANG p.K17I | ||
| ALS | Lumbosacral | M | 58 | 25 | NL | Yes | FUS p.Q210H | ||
| King et al. (2013) | FTD | Behavioural | F | 57 | ∼144 | Yes | MAPT p.A239T | ||
| ALS | FTD | Lower limb | M | 60 | ∼24 | Yes | MAPT p.A239T | ||
| ALS | FTD | Lower limb | M | 51 | ∼24 | Yes | MAPT p.A239T | ||
| van Blitterswijk et al. (2013) | FTD | F | 52 | 48 | Caucasian | Yes | GRN p.C466LfsX46 | ||
| FTD | F | 50 | >24 | Caucasian | Yes | GRN p.R493X | |||
| FTD | F | 53 | >48 | Caucasian | Yes | MAPT p.P301L | |||
| Kaivorinne et al. (2014) | FTD | Behavioural | M | 54 | >120 | Finland | Yes | TARDBP p.S292del | |
| FTD | Behavioural | F | 47 | >36 | Finland | Yes | TARDBP p.S292del | ||
| Lashley et al. (2014) | ALS | FTD | F | 59 | ∼84 | UK | Yes | GRN p.C31fs | |
| ALS | FTD | F | 46 | ∼36 | UK | Yes | GRN p.C31fs | ||
| Lattante, Le Ber et al. (2014) | FTD-ALS | Yes | ATXN2 (≥29) | ||||||
| ALS | Yes | ATXN2 (≥29) | |||||||
| Origone et al. (2015) | FTD | M | 48 | >92 | Yes | TARDBP p.N267S | |||
| Mignarri et al. (2014) | FTD | PFNA | F | 62 | >24 | Yes | GRN p.R493X | ||
| van der Zee et al. (2014) | ALS | FTD | M | 63 | 36 | Austrian | Yes | SQSTM1 p.R212C | |
| Bury et al. (2015) | ALS | FTD | F | 50 | 28 | UK | Yes | OPTN p.E322K | |
| Tarlarini et al. (2015) | ALS | FTD | Bulbar | F | 68 | 38 | Italian | Yes | FUS p.R491C |
| Pottier et al. (2015) | FTLD | agPPA | M | 68 | ∼48 | No | (1) OPTN p.Gly538Glufs*27 (2) TBK1 p.Arg117* | ||
| AD | FTLD, TDP-43 Subtype A | M | 64 | 72 | No | (1) OPTN p.Gln235* (2) OPTN p.Ala481Val | |||
| Cady et al. (2015) | ALS | Yes | (1) DCTN1 p.I196V | ||||||
| ALS | Yes | (1) SQSTM1 p.V153I | |||||||
| ALS | Yes | (1) SETX p.I2547T | |||||||
| ALS | No | (1) ATXN2 (22/31) (2) SQSTM1 p.K238E | |||||||
| ALS | No | (1) FUS p.P525L (2) ATXN2 (23/31) | |||||||
| ALS | No | (1) ATXN2 (22/32) (2) DCTN1 p.T1249I (3) SETX p.M274V |
- M, male; F, female; NL, Netherlands; UK, United Kingdom; agPPA, progressive non-fluent/agrammatic variant of primary progressive aphasia; PFNA, progressive nonfluent aphasia; & Authors suggest that this variant is benign. Blank cells are missing information
Herein, we describe two families that present with ALS and FTD. In addition to presenting with motor neuron disease, patients developed behavioral variant FTD (bvFTD). While previous reports have described such phenotypes to be related to the C9orf72 expansion, we sought to determine whether these individuals are carriers of additional variation in known ALS-FTD genes. We used an agnostic approach by initially determining the families’ C9orf72 expansion profiles in addition to sequencing other ALS-FTD associated genes. We found that both families carry a novel OPTN variant and one family has an ATXN2 expansion, and postulate that these genetic variants may be modifying the effect of C9orf72.
2 SUBJECTS AND METHODS
2.1 Study subjects and design
We identified two families carrying a C9orf72 hexanucleotide expansion, and in which several members had been assessed for the presence of neuropsychological deficits in addition to qualitative characterization of the motor system deficits. We performed genetic analysis consisting of evaluating C9orf72 and ATXN2 expansions, and targeted next generation sequencing (NGS) using the ONDRISeq, which contains probes for 80 genes associated with ALS, FTD, Alzheimer's disease, Parkinson's disease, or vascular cognitive impairment (Farhan et al., 2016). Participants were evaluated following informed consent in accordance with the Research Ethics Board at Western University. MJS conducted a neurological evaluation including the Montreal Cognitive Assessment (MoCA) on all individuals. In total, we have >20 years of documentation on the history of the families.
2.2 DNA isolation
DNA was isolated from blood collected from participants or frozen brain tissue of deceased individuals using the Gentra Puregene Blood kit (Qiagen, Venlo, Netherlands) as previously described (Farhan et al., 2016).
2.3 C9orf72
Study participants were tested for the C9orf72 expansion using: i) amplicon length analysis and ii) repeat-primed PCR. Immunohistochemistry on brain sections was performed to verify the presence of the C9orf72 dipeptide repeat proteins. Our results were validated by three research laboratories, and some samples were validated using Clinical Laboratory Improvement Amendments (CLIA) certified laboratories.
2.4 i) Amplicon length analysis and ii) repeat-primed PCR
C9orf72 G4C2 repeat was amplified using one fluorescently labeled primer followed by fragment length analysis as previously described (DeJesus-Hernandez et al., 2011; Xi et al., 2012). Allele identification and scoring was performed using GeneMapper v4.0 software. To assess the presence of an expanded C9orf72 repeat, we performed repeat-primed PCR using three primers as previously described (DeJesus-Hernandez et al., 2011; Xi et al., 2012).
2.5 Immunohistochemistry
To confirm the expansion of C9orf72 was pathogenic, we immunostained cerebellar tissue from each case for poly(GP), poly(GA), or poly(GR) proteins. Five-micron thick cerebellar slices were cut from formalin-fixed, paraffin-embedded blocks, and mounted on glass slides. After drying, slides were deparaffinized and rehydrated in xylene and alcohol washes before being steamed for 30 min in deionized water for antigen retrieval. All slides were processed on a Dako Autostainer with the Dako EnVision™+ system and 3′3-diaminobenzidine chromogen. After staining with anti-GP (Rb5823; 1:5,000), anti-GA (Rb9880, 1:50,000), or anti-GR (Rb7810, 1:2,500), slides were counterstained with Lerner hematoxylin and coverslipped with Cytoseal permanent mounting media. Cerebellar sections were examined using an Olympus BX41 microscope and images captured with cellSens Standard 1.5 software.
2.6 Southern immunoblotting
C9orf72 positive and negative DNA samples were tested via Southern immunoblotting to corroborate the repeat-primed PCR results. A digoxigenin (DIG)-labeled probe was made from gDNA of a healthy control. A total of 10 µg of gDNA was digested with XbaI at 37 °C for ∼20 hr and electrophoresed. All hybridization and washing steps were conducted as previously described (DeJesus-Hernandez et al., 2011; Xi et al., 2012). Detection of the hybridized probe DNA was performed using the CDP-star chemiluminescent substrate, and signals were visualized on LI-COR.
2.7 ATXN2 expansion testing
Study participants were screened for the ATXN2 CAG expansion using fluorescent fragment length analysis similar to the C9orf72 amplicon length analysis protocol as previously described (DeJesus-Hernandez et al., 2011; Xi et al., 2012). A laboratory with CLIA certification also validated these results.
2.8 Next generation sequencing
DNA of family 1, II-3, and family 2, III-5, were subjected to NGS. All family members were screened for any genetic variants identified in the index patients, using standard PCR and Sanger sequencing protocols.
2.9 Library preparation
Libraries were prepared using the Nextera Rapid Custom Capture Enrichment kit as previously described (Farhan et al., 2016). All samples were sequenced on the Illumina MiSeq Personal Genome Sequencer (Illumina, San Diego, CA). A total of 80 neurodegeneration specific genes were tested using ONDRISeq (Farhan et al., 2016). ALS and FTD genes tested were: ALS2, ANG, ARHGEF28, ATXN2, CENPV, CHMP2B, DAO, DCTN1, FIG4, FUS, GRN, HNRNPA1, HNRNPA2B1, MAPT, NEFH, OPTN, PFN1, PNPLA6, PRPH, SETX, SIGMAR1, SOD1, SQSTM1, TAF15, TARDBP, UBQLN2, UNC13A, VAPB, and VCP. For a full list of genes included on the ONDRISeq, see Farhan et al. (2016). It is important to mention that we did not sequence MATR3, TBK1, or NEK1, which have also been reported to be associated with ALS as the gene panel was designed and manufactured prior to the publication of these ALS gene discoveries.
2.10 Variant calling
Variant calling was performed using a customized workflow within CLC Bio Genomics Workbench v6.5 (CLC Bio, Aarhus, Denmark) as previously described (Farhan et al., 2016). Variant annotation was performed using ANNOVAR with additional databases such as the Exome Aggregation Consortium (ExAC) (Lek et al., 2016), CADD (Kircher et al., 2014), HGMD (Stenson et al., 2014), ClinVar (Landrum et al., 2016), ALS Online genetics Database (ALSoD) (Abel, Powell, Andersen, & Al-Chalabi, 2012), and our own in house databases.
2.11 Variant classification and prioritization
Variants were classified as previously described (Farhan et al., 2016). Variants were prioritized if they were rare (minor allele frequency <0.1%), exerted non-synonymous changes, were previously observed in ALS and/or FTD, and had in silico values consistent with “disease-causing” based on prediction outcomes of PolyPhen-2 (Adzhubei et al., 2010), SIFT (Kumar, Henikoff, & Ng, 2009), and CADD (Kircher et al., 2014).
2.12 Variant validation
Variants were confirmed by Sanger sequencing, as previously described (Farhan et al., 2015). Forward and reverse primers for OPTN p.Met468Arg were 5′-CTGCTATCGGAATGTACCTGG-3′ and 5′-ATGCTGATGTGAGCTCTGGG-3′, respectively. Annealing temperature for primers was 60°C.
2.13 APOE4 genotyping
Using ONDRISeq, we genotyped all individuals for the APOE risk alleles rs429358(CT) and rs7412(CT). The combination of both individual alleles determines the APOE genotype, and E4 genotype is known as a risk factor for cognitive impairment and dementia (Farhan et al., 2016).
3 RESULTS
3.1 Clinical description
Family 1 consists of individuals with multiple, distinct neurodegenerative phenotypes. Patient II-3 initially presented at age 53 with bulbar dysfunction and subsequently developed limb wasting, pyramidal weakness, and cognitive impairment. The total disease duration was 22.3 months. His brother, II-5, presents with a perplexing clinical syndrome. While some of his symptoms overlap with those of II-3, he does not meet the diagnostic criteria for ALS and has a communicating hydrocephalus, significant cervical spondolytic pathology, and a previous history of head injury from a motor vehicle accident resulting in a loss of consciousness. He is currently 70 years of age and displays progressive cognitive decline with both short and long-term memory impairment. He is unable to recall people's names and gets lost in familiar environments. He scored 10/30 on MoCA testing. He is currently wheelchair bound with significant spasticity. Their sister, II-7, was diagnosed during life with behavioral variant FTD (bvFTD) and at autopsy was found to also have motor neuron disease (MND). Their eldest sister, II-2, has no deficit on either neurological examination or on MoCA testing.
Family 2 is characterized by ALS (I-2, II-3, II-8, II-13), bvFTD and historical reports of dementia described as Alzheimer's disease (II-4 and II-17), although there are no records that would allow differentiation of Alzheimer's from FTD in these latter cases. At the age of 54, III-3 began displaying inappropriate behavior consisting of apathy, agitation, reduced verbal output, poor financial judgement, sexual indiscretion, obsessive-compulsive behavior, and repetitive movements. Within 2 years, she also developed impairment in executive function with deficits in attention and concentration, language, visuospatial construction, memory, and facial recognition. Dysarthria and dysphagia had become prominent. Her examination demonstrated ocular impersistence with preserved optokinetic nystagmus, prominent primitive reflexes, and a generalized loss of muscle bulk with diffuse, severe spasticity. Electrophysiological studies were unremarkable except for an incidental median entrapment neuropathy. Within 2 years, she had become preservative, inattentive, whispered when speaking and, in addition to diffuse motor neuron dysfunction, had mild cogwheeling. She did not consent to further electrophysiological studies, and died at age 56 years. An autopsy was not performed.
Her siblings, III-5 and III-7 had a very similar disease course. III-5, a 53-year-old accountant, first developed difficulty with organizational skills, word finding difficulties, an inability to do the daily household banking, an increased appetite, and an obsessive behavior with increasing stubbornness. Within 4 months, he had developed choking episodes, dysarthria, and reduced speech volume. His clinical and electrophysiological examinations at that point were consistent with definite ALS. This was confirmed by neuropathological examination at the time of his death 16 months following initial symptom onset, as was evidence of frontotemporal lobar degeneration. Case III-7 presented with first episode of psychosis at age 56. He began incessant smoking and became homeless. One year later he developed aspiration pneumonia and was placed on full ventilator support. His course thereafter was typical of ALS with complete anarthria, diffuse muscle wasting with fasciculations, pathologically brisk reflexes, and electrophysiological evidence of multisegmental acute and chronic motor unit remodelling. Neuropathological examination confirmed the presence of frontotemporal lobar degeneration with ALS including TDP-43 immunoreactive inclusions within cortical and spinal motor neurons.
3.2 Variants identified in patients with ALS
Patients and participating family members were screened for a hexanucleotide (G4C2) expansion in C9orf72. C9orf72 genotypes are provided in Supplementary Table S1. In families 1 and 2, we identified three and four C9orf72 expansion carriers, respectively. However, all expansions are predicted to be less than 100 repeats and within the “uncertain clinical significance” classification (Table 2, Figure 1) (Rohrer et al., 2015). These results have been confirmed independently by research and CLIA certified laboratories. In family 1, we identified three carriers (II-3, II-7, and III-6). II-3 and II-7 were diagnosed with ALS-FTD and are now deceased. The third carrier, III-6 is the son of an ALS-FTD affected individual, II-3. In family 2, we identified four C9orf72 expansion carriers (III-3, III-5, III-7, and IV-5); three carriers were diagnosed initially with bvFTD, which manifested into bvFTD-ALS. The living carrier, IV-5, is currently unaffected or presymptomatic.
| ID | Family 1 | Family 2 |
|---|---|---|
| N, affected | 2 | 8 |
| N, relatives affected with other phenotypes | 1, myelopathy; 1 Parkinson's disease | 2, Alzheimer's disease |
| Primary diagnosis | ALS | bvFTD |
| Diagnosis subcategory | FTD | ALS |
| Site of onset | Bulbar | Mixed, primarily behavioural changes |
| Female:male | 1:1 | 3:5 |
| Mean age of onset (years ± SD) | 54.4 (±2) | 55 (±8) |
| Minimum age of onset (years) | 53 | 35 |
| Maximum age of onset (years) | 56 | 64 |
| Average disease duration (months ± SD) | 29 (±7) | 25 (±8) |
| No. of C9orf72 expansion carriers | 4 | 3 |
| No. of ATXN2 intermediate length expansion carriers (22/30) | 3 | 0 |
| No. of OPTN p.Met468Arg carriers | 1 | 4 |
| APOE genotype* | E3/E2 | E3/E2 |
- * From tested individuals: family 1, II-3; and family 2, III-5. SD, standard deviation.
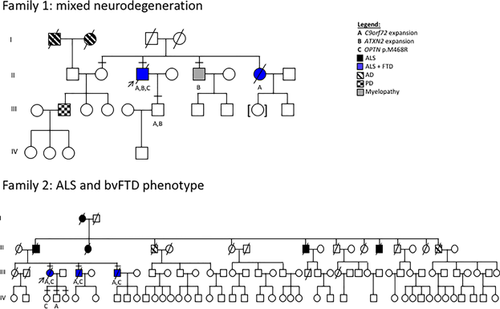
In recognition of potential C9orf72 genetic modifiers reported throughout recent literature, we screened participants for additional genetic variation. First, participants were tested for CAG expansions within ATXN2. In family 1, we observed II-3, II-5, and III-6 to be heterozygous carriers of 30 repeat units within ATXN2 (intermediate repeat length) (Table 2, Figure 1). Interestingly, the ATXN2 variation seems to segregate with spasticity as non-carriers are unaffected and are older than the typical age of onset, except for III-6, who may be presymptomatic, as he possesses both C9orf72 and ATXN2 expansions.
Concurrently, we used the ONDRISeq, to identify any additional genetic variation within neurodegeneration related genes in the families herein. In both families, we identified a novel OPTN missense variant (c.1403T>G, p.Met468Arg, rs747481280) in four individuals (family 1, II-3; family 2, III-3, III-5, and III-7), who are all are diagnosed with ALS-FTD and are C9orf72 expansion carriers; and one who is also an ATXN2 intermediate length expansion carrier (family 1, II-3) (Table 2, Figure 1, Supplementary Figure S1). Mutations in OPTN have been previously reported to be associated with ALS and FTD (Bury et al., 2015; Cirulli et al., 2015; Goldstein et al., 2016; Li et al., 2015; Maruyama et al., 2010; Pottier et al., 2015); and these variants are summarized in the ALS Online genetics Database (ALSoD), http://alsod.iop.kcl.ac.uk/misc/dataDownload.aspx#C2. We also observed the variant in IV-4, who is not a C9orf72 expansion carrier, and is currently free of any disease symptoms.
OPTN p.Met468Arg is harbored within a highly conserved ubiquitin-binding domain and is predicted to exert a damaging effect on OPTN function based on a compilation of in silico tools (Supplementary Table S2). When evaluating the frequency of OPTN p.Met468Arg in controls, we see it is absent from ExAC (n = 60,706 individuals), which is a repository of genetic information on individuals free of neurodegenerative disease. However, when using gnomAD, another much larger genetic database of 123,136 exomes and 15,496 genomes, we observe OPTN p.Met468Arg in 2 heterozygous individuals from 123,106 individuals, who have sequence coverage for this site, making the MAF = 0.0008123%. Importantly, however, gnomAD contains genetic information from individuals with a presumed non-Mendelian form of ALS, and we are unable to discern whether this OPTN variant in gnomAD is carried by these individuals from the ALS cohort or other non-ALS cohorts within gnomAD. In addition, we surveyed databases of diseased individuals, such as HGMD (n = 197,952 submissions), ClinVar (n = 442,835 submissions), and ALSoD (n = 1,096 individuals) and observed the exact variant has been observed in two individuals with ALS, and deposited in ClinVar (Black et al., 2017).
Finally, we know the inheritance of the C9orf72 expansion alone increases risk for frontotemporal dementia with cognitive impairment (Byrne et al., 2012). However, given the known association of APOE E4 carriers and the higher risk for cognitive impairment and dementia (Yu, Tan, & Hardy, 2014); and the apparent significant but modest association of the APOE E2 allele and FTD (odds ratio, 2.61; 95% confidence interval, 1.14–6.10; p = 0.03) (Chio et al., 2016), we genotyped family 1, II-3; and family 2, III-5 for APOE. We found family 1, II-3 and family 2, III-5 to carry the APOE E3, E2 genotype. The inheritance of the C9orf72 repeat expansion and other genetic variation is likely to explain much of the phenotypes observed in the patients.
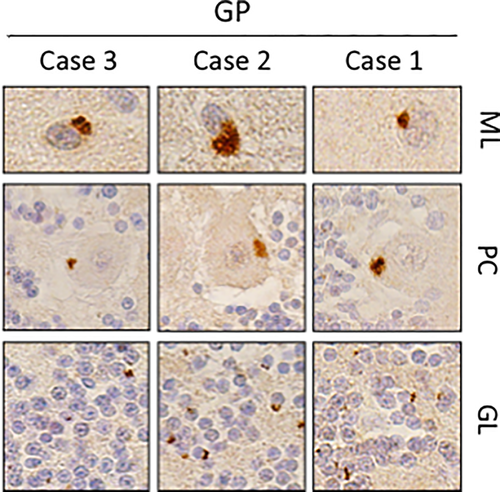
3.3 C9orf72 dipeptide immunostaining
Cerebellar sections from each case were obtained from archived neuropathological specimens and immunostained for dipeptide repeat proteins [poly(GP), poly(GA), and poly(GR)] produced from C9orf72 G4C2 expansions through repeat associated non-ATG (RAN) translation. In each instance, dipeptide repeat proteins were observed (Figure 2). These results are in keeping with previous observations that GP-positive inclusions are specific to C9orf72 repeat expansion carriers (Supplementary Figure S2) (Ash et al., 2013).
4 DISCUSSION
We have ascertained two families with multiple generations affected with ALS-FTD who are carriers of a C9orf72 expansion. Although the length of the pathological expansion falls within the equivocal range with respect to predicting disease occurrence, neuropathological examination confirmed the presence of inclusions formed of dipeptide repeat proteins within cerebellar tissue. Thus, the C9orf72 expansions reported here are pathogenic. Given the occurrence of ALS and FTD in multiple individuals, we next explored whether additional genetic variation contributed to the phenotypic heterogeneity (ALS and FTD or bvFTD). We used the ONDRISeq, a neurodegeneration specific gene panel, to sequence affected individuals and their families in addition to screening for the presence of the ATXN2 polyglutamine (polyQ) tract. We observed a rare missense variant, p.Met468Arg in OPTN, in both families. OPTN p.Met468Arg has also been previously observed in two ALS patients (Black et al., 2017), and is predicted to exert a damaging effect on the protein, however, whether this leads to loss of protein function is not clear. The variant is located within the first residue of exon 14, adjacent to the splice acceptor site therefore, it may affect splicing, leading to a loss of protein function. Previous reports have shown missense or frameshift variants in OPTN, namely, double heterozygotes: p.Gln235* and p.Ala481Val; as well as OPTN p.Gly538Glufs*27 compounded by TBK1 p.Arg117*, lead to loss of protein function in FTLD patients (Pottier et al., 2015). In our study, affected individuals carried OPTN p.Met468Arg in addition to the C9orf72 expansion, and one individual also carried an ATXN2 expansion. The mechanism of disease may be related to their convergent pathways in autophagy, where the collapse of proteostasis machinery leads to the accumulation of misfolded proteins (Bendotti et al., 2012; Cirulli et al., 2015), or altered systemic immunity resulting in neuroinflammation (Gleason, Ordureau, Gourlay, Arthur, & Cohen, 2011; Hiscott, 2007; Lall & Baloh, 2017). This is supported by the known protective role of the OPTN/TBK1 pathway, which regulate intracellular protein aggregation (Gleason et al., 2011). We conclude that the C9orf72 expansion is likely the main driver of disease, however, the burden of additional variants in other ALS genes may further modulate the phenotype.
In family 1, the index patient (II-3) carried a C9orf72 expansion, ATXN2 intermediate length expansion, and OPTN p.Met468Arg. He presented with ALS at age 53 and was found to have frontotemporal dysfunction on neuropsychological testing. His disease duration was less than 2 years. His sister's death was also due to FTD with ALS being found at autopsy. While she was documented as harbouring a C9orf72 expansion, this was done elsewhere and we were unable to obtain DNA for further analysis. II-5 carries an ATXN2 expansion and has myelopathy, spasticity, and progressive cognitive decline. The proband's son (III-6) carries both a C9orf72 expansion and an ATXN2 expansion, and is likely presymptomatic as he is currently younger than the typical age of onset within this pedigree. However, we do not know the penetrance of these variants, therefore, he may remain unaffected.
The clinical presentation was consistent in all affected individuals in family 2 in that all three patients initially presented with features of bvFTD and then progressed to either ALS or a diffuse motor neuron disease marked by prominent spasticity. Death occurred within 1–4 years following symptom onset. All affected individuals were carriers of a C9orf72 expansion as well as OPTN p.Met468Arg. Members of the subsequent generation currently do not display features suggestive of ALS or FTD, and were screened for both variants. We observed two unaffected individuals who each carried one variant, but no unaffected individual carried both variants. Based on the effect of the C9orf72 expansion, it is likely that the individual with this variant is at risk for developing ALS or bvFTD. However, whether the individual carrying only OPTN p.Met468Arg will develop ALS or bvFTD remains elusive.
In addition to our small sample size, one clear limitation of validating our findings is the inherent complexity involved in quantifying qualitative traits such as abnormal behavior or progressive cognitive decline. Much remains to be investigated regarding the pathogenicity or the genetic modifications induced by the ATXN2 expansion and the OPTN variant, however, both genes have been associated with ALS and/or ALS-FTD (Bury et al., 2015; Cirulli et al., 2015; Maruyama et al., 2010; Pottier et al., 2015; Sproviero et al., 2017; van Blitterswijk, Mullen, Heckman et al., 2014; Zhang et al., 2017). Additionally, it is important to mention that it is possible that in somatic tissues, C9orf72 repeat expansion with potentially different sizes of the pathogenic allele may contribute to the variability of the phenotype. Finally, we did not sequence MATR3, TBK1, or NEK1, which have also been reported to be associated with ALS as the gene panel was designed and manufactured prior to the publication of these ALS gene discoveries.
In conclusion, our findings highlight the importance of agnostically screening all at risk patients for all known ALS disease genes. Although today this approach remains relatively costly, recent targeted NGS approaches like the ONDRISeq facilitate these approaches on a rapid, efficient, and economical basis. Clinicians faced with patients who have atypical disease presentation are encouraged to consider screening for additional mutations in related genes to identify possible oligogenic interactions. By identifying additional genetic variation, complex phenotypes can be explained by the additive effect of variants with large or small, yet significant effects that converge on the same disease pathway. In addition, screening of multiple genes is important for the genetic counseling of family members who are carriers of one but not all pathogenic variants.
ACKNOWLEDGMENTS
We are grateful to the participating families in our study and to Mrs. Ann Rowe for assistance in the clinical and genotyping of the pedigrees. Many thanks to Dr. Ming Zhang and Dr. Ekaterina Rogaeva from the Tanz Centre for Research in Neurodegenerative Diseases at the University of Toronto for experimental guidance and validation. Thank you to Dr. Kathryn Volkening for technical assistance during the Southern blot experiments. Thank you to Ms. Monica Castanedes-Casey and Dr. Dennis Dickson from the Department of Neurology at the Mayo Clinic, Jacksonville, Florida for assistance with the immunohistochemistry assays. This research was supported by the Canadian Institutes of Health Research (CIHR), ALS Canada, and the McFeat family fund. RAH is supported by CIHR, Heart and Stroke Foundation, and Genome Canada. SMKF is supported by the CIHR Fredrick Banting and Charles Best Canada Graduate Scholarship.
CONFLICT OF INTEREST
None.