Suggestive evidence for linkage of ADHD features in bipolar disorder to chromosome 10p14†
How to Cite this Article: Joo E-J, Greenwood TA, Schork N, McKinney RA, Sadovnick AD, Remick RA, Keck PE, McElroy SL, Kelsoe JR. 2010. Suggestive Evidence for Linkage of ADHD Features in Bipolar Disorder to Chromosome 10p14. Am J Med Genet Part B 153B:260–268.
Abstract
Higher rates of bipolar disorder amongst the first-degree relatives of probands with ADHD, and increased rates of ADHD in the relatives of bipolar probands have been reported in many studies. This suggests some commonality in the genetic basis for bipolar disorder and ADHD. We hypothesized that ADHD symptoms in bipolar disorder may access a quantitative subphenotype that is genetically less complex and therefore advantageous for mapping studies. The Wender Utah Rating Scale (WURS) was used to quantify ADHD features in 57 bipolar families collected for linkage studies. The factor structure of the WURS was first examined, and heritability was estimated. Linkage analysis was then conducted using the WURS total score and factor scores as quantitative traits. Three factors were identified: impulsivity and defiant behavior, mood instability and anxiety, and inattention. The total WURS and factor scores were each significantly heritable (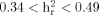 ) in bipolar families. The inattention factor obtained maximum evidence of linkage on chromosome 10p14 (LOD = 3.35, 25 cM). A LOD score of 2.06 for the total WURS score was found on chromosome 12q24 region. Childhood ADHD features in patients with bipolar disorder are heritable and may represent a genetically distinct dimension of illness. 10p14, in particular may contain a locus for inattention in bipolar disorder. Quantitative dimensional phenotypes such as this may be useful for both mapping of genes and understanding the role of genes in bipolar disorder. © 2009 Wiley-Liss, Inc.
) in bipolar families. The inattention factor obtained maximum evidence of linkage on chromosome 10p14 (LOD = 3.35, 25 cM). A LOD score of 2.06 for the total WURS score was found on chromosome 12q24 region. Childhood ADHD features in patients with bipolar disorder are heritable and may represent a genetically distinct dimension of illness. 10p14, in particular may contain a locus for inattention in bipolar disorder. Quantitative dimensional phenotypes such as this may be useful for both mapping of genes and understanding the role of genes in bipolar disorder. © 2009 Wiley-Liss, Inc.
INTRODUCTION
Family, twin, and adoption studies have consistently indicated that bipolar disorder has a strong genetic component [Taylor et al., 2002]. However, identifying specific genes for bipolar disorder has been challenging. To be successful in finding genes, it is likely important to use a genetically relevant definition of phenotype. A phenotype with high heritability and low heterogeneity would be ideal [Schulze et al., 2006]. Patients with bipolar disorder display significant clinical variability such as: psychotic features, suicidality, rapid cycling, substance use, panic attacks, and others. This may reflect underlying heterogeneity at the biological and genetic levels. Therefore, current DSM diagnosis as a phenotype probably does not have the best power to detect genes for such a heterogeneous trait. In this situation, if an intermediate phenotype can subdivide the affected subjects into biologically more homogeneous groups, it may be a useful alternative for mapping genes for a complex trait like bipolar disorder. Further, a quantitative phenotype usually has better statistical power than a qualitative categorical phenotype, especially for a trait with multiple genes [Wijsman and Amos, 1997]. As more evidence supports a polygenic etiology for bipolar disorder [Kelsoe, 2003; Baum et al., 2007], a genetic approach with a biologically relevant quantitative subphenotype could be more promising for bipolar disorder.
Diagnostic subgroups, clinical features and various comorbid conditions of bipolar disorder have been considered as possibly valuable alternative phenotypes. Recent epidemiological studies have shown that comorbid conditions such as substance abuse, alcoholism, and psychosis, as well as, clinical features such as suicidal attempts and social functioning were strongly familial [MacKinnon et al., 2005; Schulze et al., 2006]. Even though it is not clear that these clinical features and various comorbid conditions are genetically less complex, it is possible that more homogenous groups of patients can be selected by applying these variables. Attention Deficit Hyperactivity Disorder (ADHD) is one of the most frequent comorbid psychiatric conditions among bipolar patients, but the comorbid ADHD in bipolar disorder has not yet been studied for heritability or linkage.
Epidemiological studies in the US have shown that ADHD co-occurs in up to 85% of children with bipolar disorder and bipolar disorder exists in up to 22% of children with ADHD [Althoff et al., 2005; Singh et al., 2006]. In addition to this remarkably high comorbidity between ADHD and bipolar disorder, higher rates of bipolar disorder are also found amongst the first-degree relatives of probands with ADHD, and increased rates of ADHD have been reported in the relatives of bipolar probands. One meta-analysis showed that ADHD is three times more common in the relatives of probands with bipolar disorder than normal controls, and bipolar disorder is twice as common in relatives of probands with ADHD than normal controls [Faraone et al., 1997]. These studies suggest several possibilities as below. Both disorders share some common genetic susceptibility. ADHD features could be a reasonable trait showing genetic overlap with bipolar disorder. It is also possible that ADHD features in bipolar disorder may derive from a distinct subset of genes and be a useful subphenotype for mapping studies. Clinically and diagnostically, ADHD features are already confounded with bipolar disorder. Therefore, some cases must be ambiguous in diagnosis, especially in childhood.
The Wender Utah Rating Scale (WURS) was developed to measure retrospectively the severity of ADHD symptoms of subjects in childhood. It is a self-report questionnaire developed originally with 61 items in order to identify ADHD in childhood. The WURS has been shown to be sensitive in detecting ADHD [Ward et al., 1993; McCann et al., 2000] and to have good internal consistency reliability and test-retest reliability [Ward et al., 1993; Fossati et al., 2001]. Several studies have found a similar three-factor structure for the scale. McCann et al. [2000] studied 143 adults referred to an adult ADHD specialty clinic and found three factors: dysthymia, oppositional/defiant behavior, and school problems. Fossati et al. [2001] studied three independent samples (college students, psychiatric patients, and secondary school students) with an Italian version of WURS and found three factors: hyperactivity/irritability, affective symptoms, and inattention.
Here we describe our examination of the WURS in bipolar families. We used the WURS to measure ADHD features in childhood and found three factors consistent with previous studies. Then we calculated heritabilities for the total WURS score, as well as each of the three factors. We found that the WURS scores (total score and three factor scores) in bipolar families are significantly heritable in our sample. Therefore, we hypothesized that ADHD symptoms in bipolar disorder may access a quantitative subphenotype that is genetically less complex and, therefore, advantageous for mapping studies. We conducted linkage analysis using the WURS scores as phenotypes and found a suggestive linkage peak at chromosome 10 for the inattention factor.
METHODS
Subjects
Subjects with bipolar disorder and their family members were derived from three different data sets collected for genetic studies of bipolar disorder. One data set (University of California San Diego, UCSD) was recruited from one of three sites (San Diego, Vancouver, and Cincinnati) as part of a collaborative genetic linkage study of bipolar disorder [Kelsoe et al., 2001]. The other two data sets were collected in San Diego as part of the National Institute of Mental Health (NIMH) Genetics Initiative for Bipolar Disorder Waves 3 and 4 [Dick et al., 2003]. WURS was collected only at the San Diego site of the NIMH consortium, so data was not available for the rest of this collection. An independent set of 257 sporadic (i.e., nonfamilial) bipolar cases with WURS data collected in San Diego was included in the factor analyses. Normal controls were collected by advertising in the UCSD Mental Health Clinical Research Center and screened using the SCID as below for the absence of psychiatric illness. All subjects were Caucasians of European ancestry.
Written informed consent was obtained using procedures approved by each local university IRB. Families were first identified through a proband diagnosed with bipolar I disorder or bipolar II disorder (UCSD sample) or a bipolar I sib pair (Waves 3 and 4) [Dick et al., 2003]. Each subject was interviewed and diagnosed using either a modified version of the Structured Clinical Interview for DSM-III-R (SCID) [Spitzer et al., 1992] or using the Diagnostic Interview for Genetics Studies (DIGS) [Nurnberger et al., 1994]. Interviewers were extensively trained and reliability regularly tested. A panel of clinicians, blind to genotype, reviewed the interview, medical records, and information from available family members to make the final DSM-IV diagnosis.
Factor analyses were conducted on 540 subjects from this sample that included all phenotyped subjects in the 57 bipolar families (N = 227), the sporadic cases with bipolar disorder (N = 257), and the normal controls (N = 56). Heritability and linkage analyses were performed in the 57 bipolar families, which included 316 subjects, almost all of which were collected in San Diego (Table I) and an average of 5.5 individuals per family, including probands, affected and unaffected siblings, parents, and grandparents. There were 129 sib-pairs, 5 half-sib pairs, 350 parent-child pairs, 96 grandparent-grandchild pairs, 8 cousin pairs, and 68 avuncular pairs. For the sake of computation efficiency, these families were pruned to remove more distantly related individuals that did not significantly contribute to the information content of the family. As a result, among the 227 subjects from bipolar families included in the factor analyses, 9 subjects with WURS were removed.
| UCSD | Wave 3 | Wave 4 | Combined | |
|---|---|---|---|---|
| Families | 26 | 9 | 22 | 57 |
| Total subjects | 173 | 35 | 108 | 316 |
| Subjects with genotypes and WURS | 123 | 24 | 69 | 216 |
| Subjects with genotypes | 138 | 28 | 80 | 246 |
| Subjects with WURS | 124 | 25 | 69 | 218 |
| Informative pairs | 293 | 21 | 83 | 397 |
| Markers genotyped | 436 | 372 | 398 | 1,206 |
Measurement of ADHD Features
We used a modified short form of WURS to retrospectively measure the severity of ADHD symptoms of subjects in childhood. This is a self rated scale including 22 items of childhood ADHD symptoms that are each rated on a 5-point scale (0–4).
Genotype Data
Each of the three family sets, UCSD, Wave 3 and Wave 4, were separately genotyped as part of independent linkage genome scans of bipolar disorder [Kelsoe et al., 2001; Dick et al., 2003]. Though the genotyping methods were comparable, somewhat different microsatellite sets were used. Microsatellite genotyping for the UCSD sample was performed at UCSD, while waves 3 and 4 were genotyped at the Center for Inherited Disease Research. The markers for each dataset were primarily tri- and tetranucleotide repeats. Each was genotyped using multiplexed methods and detected using fluorescent tags as described previously [Kelsoe et al., 2001; Dick et al., 2003]. A total of 436 microsatellite markers were genotyped with an average intermarker interval of 8 cM for UCSD sample. The wave 3 and wave 4 samples included 372 and 398 markers respectively. Two hundred twenty seven markers overlapped among three data sets.
Statistical Methods
Factor analysis
A principal components analysis was performed to determine the factor structure. The Kaiser–Meyer–Olkin measure of sampling adequacy was 0.94, and the significance of Barlett's test of sphericity was <0.001 for this sample. These figures supported the factorability of the correlation matrix of this sample. Primary factor analysis was done without rotation and found three related components with Eigen values greater than 1.0. An inspection of the screen plot revealed a break after the three components. Since the three factors were correlated each other, promax rotation with Kaiser normalization was conducted to maximize the loading of each variable on one of the extracted factors and to minimize the loading on all other factors [Field, 2000]. Rotation converged in 6 iterations. SPSS 12.0 was used for this analysis.
ANOVA test and correlation test
Our sample for factor analysis could be subdivided into four different diagnostic groups: 104 patients with bipolar disorder, 37 patients with major depressive disorder, 30 unaffected relatives, and 56 unrelated healthy controls. We analyzed WURS scores between diagnostic groups using ANOVA. Post hoc comparisons were performed with the Tukey HSD. We also conducted a Pearson correlation test to investigate any correlation between variables (all WURS scores and age at interview) by using SPSS 12.0.
Heritability analysis
We examined heritability for the total WURS score, as well as for individual factor scores, in 316 subjects from 57 bipolar pedigrees. Heritability (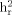 ) estimates were obtained via the variance component methodology implemented in the Sequential Oligogenic Linkage Analysis Routines (SOLAR) v.2.1.2 package [Almasy and Blangero, 1998]. This maximum likelihood method assumes a multivariate normal distribution of phenotypes in a pedigree and can accommodate a defined set of covariates. The null hypothesis of no heritability (
) estimates were obtained via the variance component methodology implemented in the Sequential Oligogenic Linkage Analysis Routines (SOLAR) v.2.1.2 package [Almasy and Blangero, 1998]. This maximum likelihood method assumes a multivariate normal distribution of phenotypes in a pedigree and can accommodate a defined set of covariates. The null hypothesis of no heritability (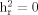 ) is tested by comparing a “full” model, which assumes that some fraction of the phenotypic variation is explained by genetic factors, to a “reduced” model, which assumes that no variation is explained by genes, using likelihood ratio tests. Covariates (age and sex) that were significant in the heritability analysis were retained and considered in the linkage analyses.
) is tested by comparing a “full” model, which assumes that some fraction of the phenotypic variation is explained by genetic factors, to a “reduced” model, which assumes that no variation is explained by genes, using likelihood ratio tests. Covariates (age and sex) that were significant in the heritability analysis were retained and considered in the linkage analyses.
Linkage analysis
A combined dataset from three sets of genotype data was used for analysis. Different but overlapping sets of microsatellite markers were used in the three genome scans. In order to accommodate the difference in markers used in each dataset, and possible differences in allele coding for the same markers used across samples, we created a combined dataset for analysis using a similar method of combining data as described by McQueen et al. [2005]. All markers from each dataset were included in one map. Markers available only in one sample were coded as missing data in the other. Markers used in both samples were coded as two different markers at the same map position, in order to accommodate possible differences in allele coding. The resulting genotype dataset was evaluated using the pedstats module of Merlin [Abecasis et al., 2002] to check the pedigrees for non-Mendelian inheritance. Marker order and map positions were obtained from a modified version of the deCODE genetic map [Nievergelt et al., 2004].
Although there are many different methodologies that can be used to assess linkage for quantitative traits, we took advantage of the regression-based method implemented in the software Merlin-regress of the Merlin linkage analysis package [Sham et al., 2002; Nievergelt et al., 2004]. Merlin-regress has been shown to be more robust to issues involving incomplete marker informativity and is appropriate for selected sample [Cordell, 2004; Schork and Greenwood, 2004a,b; Franke and Ziegler, 2005]. Missing genotypes are imputed and assessed probabilistically by conditioning on all other linked marker data and pedigree structure. The proportion of marker alleles shared identical-by-descent (IBD) among all relative pairs is estimated independently for all autosomal markers. For multipoint quantitative trait linkage analyses, LOD scores and P values were computed at 1 cM intervals along each chromosome starting at the first marker. Age at interview and sex were also included as covariates in the analysis as appropriate.
Merlin was also used to conduct simulation analyses to estimate the empirical significance of the highest linkage peaks in the study. This method involves gene dropping simulations to replace the input data with simulated chromosomes that maintain the family structure, marker spacing, allele frequencies, and missing data patterns present in the original study. For each LOD score >2.0, 1,000–10,000 simulations of the entire chromosomes were performed, and regression linkage analyses were run for each of these simulated data sets. Using the traditional method, we recorded the fraction of times a simulated LOD score exceeded the actual LOD score and divided by the number of simulations performed.
RESULTS
Factor Structure
The principal components analysis of the modified short form of WURS with promax rotation found three related factors: impulsivity and defiant behavior (IMP), mood instability and anxiety (MOOD), and inattention (INATT). These three factors accounted for 62.82% of the variance, with the IMP factor contributing 47.08%, the MOOD factor contributing 8.77%, and the INATT factor contributing 7.97%. Three factors were significantly correlated to each other (correlation coefficient > 0.3). Table II summarizes the results of the factor analysis.
| Factor 1, IMP | Factor 2, MOOD | Factor 3, INATT | |
|---|---|---|---|
| WURS items | |||
| Temper outbursts, tantrum | 0.93 | −0.03 | −0.18 |
| Disobedient, rebellious, sassy | 0.91 | −0.15 | 0.01 |
| Hot- or short-tempered, low boiling point | 0.82 | 0.07 | −0.12 |
| Stubborn, strong-willed | 0.75 | −0.07 | −0.09 |
| Trouble with authorities and school, visits to principal's office | 0.73 | −0.29 | 0.26 |
| Tendency to be or act irrational | 0.63 | 0.06 | 0.20 |
| Acting without thinking, impulsive | 0.63 | −0.02 | 0.29 |
| Angry | 0.54 | 0.53 | −0.17 |
| Trouble seeing things from someone else's point of view | 0.45 | 0.17 | 0.21 |
| Anxious, worrying | −0.28 | 1.00 | −0.01 |
| Sad or blue, depressed, unhappy | 0.01 | 0.90 | −0.10 |
| Low opinion of myself | −0.14 | 0.84 | 0.11 |
| Nervous, fidgety | −0.09 | 0.81 | 0.07 |
| Moody, ups, and downs | 0.26 | 0.69 | −0.02 |
| Irritable | 0.45 | 0.59 | −0.14 |
| Unpopular with other children | 0.01 | 0.45 | 0.26 |
| Overall a poor student, slow learner | −0.07 | −0.10 | 0.85 |
| Not achieving up to potential | 0.02 | −0.05 | 0.85 |
| Trouble with mathematics or numbers | −0.09 | −0.05 | 0.75 |
| Concentration problems, easily distracted | 0.00 | 0.25 | 0.62 |
| Trouble with stick-to-it-tiveness | 0.10 | 0.23 | 0.57 |
| Inattentive, daydreaming | 0.08 | 0.27 | 0.54 |
| Initial Eigen values | 10.14 | 1.93 | 1.75 |
| Eigen values after rotation | 8.28 | 8.17 | 6.60 |
| % of variance | 46.08 | 8.77 | 7.97 |
| Component correlation | |||
| IMP | 1.00 | 0.62 | 0.53 |
| MOOD | 0.62 | 1.00 | 0.53 |
| INATT | 0.53 | 0.53 | 1.00 |
- WURS, Wender Utah Rating Scale; IMP, impulsivity and defiant behavior, MOOD, mood instability and anxiety; INATT, inattention.
Descriptive Statistics, Comparison Between Diagnostic Groups and Correlations
The mean ages at interview for each diagnostic group were 41.39 for bipolar disorder, 47.48 for major depressive disorder, and 48.23 for unaffected relatives in bipolar families (F = 3.90, df = 2, P = 0.02). WURS total and factor scores for each diagnostic group are illustrated in Figure 1. Total and factor scores of WURS were highest in bipolar subjects, followed by subjects with major depressive disorder, unaffected relatives in bipolar families, and then normal controls. The overall differences in total and factor scores of WURS across diagnostic groups were statistically significant (ANOVA, Total score: F = 40.88, df = 3, P < 0.001, IMP: F = 17.97, df = 3, P < 0.001, MOOD; F = 50.59, df = 3, P < 0.001, INATT: F = 16.62, df = 3, P < 0.001). Post hoc analysis found that for WURS total score, there was a significant difference between most pairs of diagnostic groups. For IMP, the bipolar disorder group differed from other diagnostic groups, but major depressive disorder group was not significantly differed from unaffected relatives or controls. For MOOD and INATT, there was no significant difference in scores between bipolar disorder and major depressive disorder groups. Unaffected relatives and normal controls were not significantly different for WURS total or any of the three factors (Table III). All WURS scores (total and factor scores) were significantly correlated with age at interview (total r = −0.29; IMP, r = −0.41; MOOD r = −0.21; all P < 0.001). This correlation was not as strong for INATT (r = −0.14, P = 0.02).

WURS total and factor scores between diagnostic groups. ANOVA tests were performed to compare diagnostic groups for total and factor scores of WURS. IMP, impulsivity and defiant behavior; MOOD, affective instability and anxiety; INATT, inattention; BP, bipolar disorder I; bipolar disorder II, and Schizoaffective disorder; MDD, major depressive disorder; REL, unaffected relatives; HC, healthy controls.
| Comparing Groups | Post hoc P-values | ||||
|---|---|---|---|---|---|
| I | II | Total | IMP | MOOD | INATT |
| BP | MDD | 0.03 | 0.01 | 0.08 | 0.95 |
| BP | REL | 0.00 | 0.00 | 0.00 | 0.00 |
| BP | HC | 0.00 | 0.00 | 0.00 | 0.00 |
| MDD | REL | 0.02 | 0.97 | 0.00 | 0.02 |
| MDD | HC | 0.00 | 0.06 | 0.00 | 0.00 |
| REL | HC | 0.15 | 0.23 | 0.32 | 0.75 |
- Post hoc comparison by Tukey HSD.
- BP, bipolar disorder I, bipolar disorder II, Schizoaffective disorder; MDD, major depressive disorder; REL, unaffected relatives; HC, healthy controls; IMP, impulsivity and defiant behavior; MOOD, mood instability and anxiety; INATT, inattention.
Heritability of WURS Scores
Heritability measures were highly significant for total WURS and all factor scores (Table IV). WURS total score was highly heritable (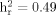 ) and more heritable than any individual factor. Since age at interview was found to be a significant covariate for total WURS and all three factors, it was included as a covariate in the subsequent linkage analyses. Sex was a significant covariate for IMP and INATT and was also included as a covariate in the linkage analyses for these factors. The proportion of variance due to all final significant covariates ranged from 6% (INATT) to 23% (IMP). Residual kurtosis values were in normal range for all WURS scores.
) and more heritable than any individual factor. Since age at interview was found to be a significant covariate for total WURS and all three factors, it was included as a covariate in the subsequent linkage analyses. Sex was a significant covariate for IMP and INATT and was also included as a covariate in the linkage analyses for these factors. The proportion of variance due to all final significant covariates ranged from 6% (INATT) to 23% (IMP). Residual kurtosis values were in normal range for all WURS scores.
| Trait | 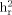 (SE) (SE) |
P-value | P-value | |
|---|---|---|---|---|
| Age | Sex | |||
| Total | 0.49 (0.17) | 0.00 | 0.00 | 0.15 |
| IMP | 0.41 (0.16) | 0.00 | 0.00 | 0.00 |
| MOOD | 0.34 (0.15) | 0.01 | 0.00 | 0.68 |
| INATT | 0.41 (0.16) | 0.00 | 0.03 | 0.04 |
-
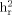 (SE), heritability (standard error).
(SE), heritability (standard error).
- All values were calculated by SOLAR.
- Age: P-value of age as a covariate.
- Sex: P-value of sex as a covariate.
- WURS, Wender Utah Rating Scale; IMP, impulsivity and defiant behavior; MOOD, mood instability and anxiety; INATT, inattention.
Linkage Analysis of WURS Scores
Table V and Figure 2 summarize the prominent results of the linkage analyses using Merlin-regress. The maximum evidence for linkage was found on chromosome 10p14 for INATT with a LOD of 3.35 at 25 cM. The simulation analyses yielded an empirical P-value of 0.0002 for this locus, providing further evidence to support linkage to this region. The second best peak was found on chromosome 12q24 for the total WURS score with a LOD of 2.06 at 161 cM. The simulation analyses yielded an empirical p-value of 0.001 for this locus. There were two more linkage peaks for WURS factor scores with LOD score greater than 1.5: IMP on chromosome 3 (LOD = 1.79, 82 cM) and INATT on chromosome 8 (LOD = 1.78, 39 cM). Analyses of the MOOD factor did not reveal a LOD score >1.5 for any chromosomal region.
| Trait | Chromosome | Position (cM) | LOD | Peak width |
|---|---|---|---|---|
| Total | 10 | 17 | 1.49 | 11–24 |
| 12 | 161 | 2.06 | 151–168 | |
| 15 | 127 | 1.26 | 124–131 | |
| IMP | 1 | 271 | 1.08 | 266–275 |
| 2 | 153 | 1.21 | 149–157 | |
| 3 | 82 | 1.79 | 72–87 | |
| 12 | 160 | 1.33 | 156–165 | |
| 15 | 124 | 1.09 | 122–128 | |
| MOOD | 2 | 42 | 1.25 | 35–47 |
| 5 | 10 | 1.01 | 10 | |
| 10 | 20 | 1.05 | 19–20 | |
| 12 | 161 | 1.06 | 159–162 | |
| INATT | 1 | 275 | 1.40 | 266–275 |
| 5 | 0 | 1.33 | 0–1 | |
| 8 | 39 | 1.78 | 33–47 | |
| 10 | 25 | 3.35 | 9–30 | |
| 15 | 129 | 1.01 | 128–129 |
- WURS, Wender Utah Rating Scale; IMP, impulsivity and defiant behavior; MOOD, mood instability and anxiety; INATT, inattention.
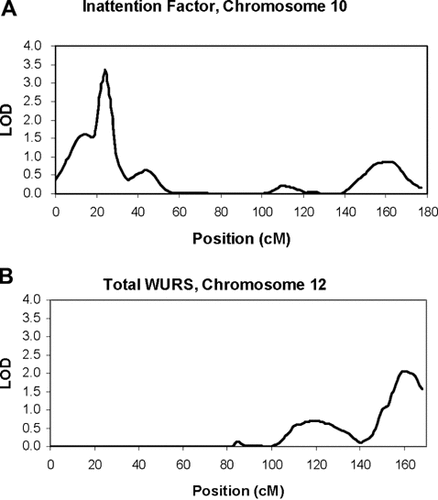
Multipoint linkage analyses (LOD > 2.0). The multipoint linkage results of two linkage regions identified: (a) Linkage curve of inattention factor score of WURS on chromosome 10; (b) Linkage curve of total score of WURS on chromosome 12.
DISCUSSION
Our analysis showed that WURS displayed a three-factor structure similar to that shown in other studies. Total and factor scores of WURS were significantly different between diagnostic groups and highly heritable. We found a suggestive linkage peak for INATT on chromosome 10p14 and another linkage peak on chromosome 12q24 for total WURS score.
Several limitations of this study must be considered in order to interpret our results. We used the WURS to measure childhood ADHD symptoms. This is a subjective self-rated scale, and thus may have biases related to the subject's subjective evaluation of childhood emotion and behavior retrospectively. More importantly, scoring may be influenced by recall bias due to aging, or possibly a cohort effect. Consistent with a recall or cohort effect, we found a significant effect of age at interview on total and factor scores of WURS, thus we included this variable as a covariate in our model for further analyses of heritability and linkage. It was noticeable that the effect of age at interview was less strongly correlated with INATT. This could indicate INATT is relatively less influenced by recall bias due to aging. Another important limitation is limited statistical power due to insufficient sample size. In this study, we included 57 families, 316 individuals as a whole. However, both WURS and genotype information was not available for large proportion of our family members (available only for 216 subjects). This restricted sample size reduced the advantage of adapting quantitative subphenotype. Additionally, genotype data from three different data sets could not be combined directly because of possible inconsistency in allele calling. This reduced the statistical power of combining samples in this study.
The three factors we found in our sample were consistent with the factor structure found in previous studies [McCann et al., 2000; Fossati et al., 2001]. Our sample included mostly bipolar patients and relatives, as well as a relatively small number of controls, in comparison to pervious studies that were done with subjects with ADHD or normal controls. It suggests that the WURS measures three phenotypic domains of ADHD features: impulsivity, mood instability, and inattention, regardless of the diagnosis of subject. Total and factor scores of the WURS were different between diagnostic groups. Bipolar disorder differed from relatives and controls for all three factors, in contrast to major depressive disorder, which differed between relatives and controls only for the MOOD and INATT, but not for the IMP. Major depressive disorder differed from bipolar disorder for total WURS as well as the IMP, but not for MOOD or INATT. This suggests that bipolar disorder and major depressive disorder share some symptoms in childhood, but the clinical manifestations do not overlap completely. Some of these symptoms also overlap with ADHD as part of either a prodrome of mood disorder or comorbidity with ADHD. Childhood impulsivity could be a differential manifestation of bipolar disorder distinguishing it from major depressive disorder. This indicates that the impulsivity factor could be a useful tool to predict long term polarity of affective disorders, as has been suggested previously [Swann et al., 2007]. Our analyses could not find a significant difference between unaffected relatives of bipolar disorder patients and healthy controls, but there was a trend. Such a difference would be ideal for an endophenotype, but it is possible that the difference could not be detected because of limited number of healthy controls in our sample.
Chromosome 10p14 has shown linkage for both bipolar disorder and schizophrenia. Researchers found a suggestive LOD score in US families with bipolar disorder from the NIMH Genetics Initiative sample [Rice et al., 1997; Foroud et al., 2000]. The two highest linkage spots with the model of bipolar disorder type I and type II in these studies were identical (54 and 115 cM on chromosome 10). Our highest linkage spot was not same with these peaks, ours was more proximal at 25 cM on chromosome 10. This indicates our linkage finding for inattention factor is not just another finding of the same linkage of bipolar disorder. More recent stage 2 of the Wellcome Trust UK-Irish bipolar affective disorder sibling-pair genome screen study found some evidence for linkage at two spots on chromosome 10 [Lambert et al., 2005]. One is at 29 cM, which is near to our linkage peak for inattention. The other is at 45 cM, which is close to previous linkage studies on bipolar disorder from the NIMH Genetics Initiative sample. This also supports that on chromosome 10, there maybe at least two linkage peaks with bipolar disorder and more analyses are required to dissect this complex genetic feature. We assume that more homogenous subphenotype could provide a key to resolve the complexity. Interestingly, there have been a linkage and an association studies supporting this chromosomal region for susceptibility locus of schizophrenia [Schwab et al., 1998, 2006]. Our multipoint linkage peak for the inattention factor was found on 10p14 region with LOD score of 3.35, which was supported by simulations to assess empirical significance (empirical P-value = 0.0002). This would be considered as a significant linkage (LOD > 3.3) by Lander and Kruglyak [1995]. However, if we consider our 4 genome scans as separate 4 genome scans, even though the phenotypes are not completely independent to each other, the significance level should be adjusted for multiple testing. Then this finding would be better considered as a suggestive linkage. To our knowledge, 10p14 has not been implicated for ADHD. However, it is possible that a common susceptibility locus for bipolar disorder, schizophrenia, and childhood inattention might be located in this region. This chromosomal region is a relative gene desert and contains only several genes for hypothetical proteins for which little information is available. There are no obvious nearby candidate genes.
We found a linkage peak on chromosome 12q24 for total WURS score in this sample. The LOD score 2.06 was not suggestive for linkage according to Lander and Kruglyak, although the simulations supported the finding with an empirical P-value of 0.001. Supportive evidence of linkage on chromosome 12q24 region with bipolar disorder has been repeatedly found in many studies of various populations, such as French Canadian isolate [Morissette et al., 1999; Shink et al., 2005], Faroe islands isolate [Degn et al., 2001], United Kingdom and Iceland [Curtis et al., 2003], Irish population [Cassidy et al., 2007] and two Danish families sample [Ewald et al., 1998, 2002]. The highest peaks in the Ewald et al. [2002] and Shink et al. [2005] studies were at 148 cM (near D12S1639) and 142 cM (near D12S378), respectively, both of which are only 14–20 cM proximal to our peak at 162 cM (near D12S367). Significant associations between bipolar disorder with polymorphic markers within candidate genes, such as Citron [Lyons-Warren et al., 2005], Slynar [Kalsi et al., 2006], and DAO [Prata et al., 2007], have been found observed for this chromosomal region also. Recently a non-conservative amino acid change from a glutamine to arginine in the P2RX7 gene, encoding a central nervous system-expressed purinergic receptor, also has been reported to be significantly associated with both bipolar and unipolar affective disorders [McQuillin et al., 2009]. There was a study reported association between this chromosomal region and schizophrenia [Reif et al., 2006]. In addition to the linkage signal total WURS score, more modest evidence to suggest linkage to this region was observed for the IMP and MOOD factor scores. Intriguingly, the INATT factor score did not reveal linkage to this. This phenomenon may suggest that the IMP and MOOD factors of the WURS may be more representative of bipolar disorder, whereas the INATT factor could be a more unique feature of ADHD. To our knowledge, the chromosome 12q24 region has not been suggested for ADHD itself.
Family studies have consistently found co-occurrence or bidirectional overlap between bipolar disorder and ADHD [Faraone et al., 1997; Althoff et al., 2005]. This suggests that they may share aspects of underlying pathophysiology, and a common genetic susceptibility. Hypotheses to explain this co-occurrence between ADHD and bipolar disorder have been suggested by various researchers. Faraone et al. [1997] examined the patterns of comorbidity between bipolar disorder and ADHD in a series of family studies, and suggested a familial and genetic etiology for this co-occurrence [Althoff et al., 2005]. These data suggested that bipolar disorder with childhood ADHD features might represent a distinct form of illness. Our study result suggests further hypotheses about the relationships between childhood ADHD features and bipolar disorder. First, ADHD features in childhood could identify a genetically more homogenous subgroup among bipolar families. This would be consistent with the idea of Faraone et al. that the co-occurrence of both disorders represents a distinct form of illness. Another possibility is that the putative loci we have identified may play a role in both disorders, and represent a shared genetic basis. A third possibility is that these loci may have little effect on risk for bipolar disorder, but act as modifiers causing childhood onset of ADHD features. It is also possible, that these loci simply are associated with early onset of bipolar disorder, which shares some clinical features, but not a genetic basis with ADHD. To explore the latter possibility, we performed an additional linkage analysis using age at onset of mood disorders as a quantitative phenotype. We failed to find evidence for significant or suggestive linkage to any chromosomal region, including 10p14 or 12q24, in these families (data not presented), which refutes the hypothesis that our results are merely associated with an early onset form of bipolar disorder. Ultimately, the identification of specific genes for both disorders and for co-morbid families will be necessary to resolve these possible scenarios.
It is also striking that the factor with the highest LOD score was INATT on 10p14. INATT explains only 8% of the variance of the WURS, yet proved to be quite powerful as a quantitative phenotype. This suggests that the INATT factor may be influenced by a distinct subset of genes and a valuable way to reduce heterogeneity. This illustrates the utility of such quantitative traits for mapping. However, the small variance observed for the inattention factor could be considered as a limitation of this study.
In conclusion, our factor analysis revealed three correlated factors in the WURS, and the total score and all three factor scores showed strong heritability. Linkage analyses indicated a suggestive a linkage of 10p14 to INATT and 12q24 to total WURS score in these bipolar families. These chromosomal regions have been previously implicated in bipolar disorder and schizophrenia in different samples. Our results suggest that specific chromosomal loci may be linked to ADHD features in bipolar disorder. This may point to specific genes that mediate early ADHD symptoms in bipolar disorder, or possibly shared between bipolar disorder and ADHD. Therefore, our findings may be relevant to the genetics of ADHD, bipolar disorder, or a form of illness with features of both ADHD and bipolar disorder. These results also suggest that ADHD features measured by the WURS may be a useful quantitative subphenotype to find genes for bipolar disorder. Further genetic studies with intermediate phenotypes are necessary to facilitate finding genes for complex disorder such as bipolar disorder.
Acknowledgements
We would like to thank the family members who participated in this study. This work was supported by Novartis Parma AG, and by grants to J.R.K. from the Department of Veterans Affairs and the National Institute of Mental Health (NIMH) (MH47612, MH59567, MH68503), the UCSD Mental Health Clinical Research Center (MH30914), the UCSD General Clinical Research Center (M01 RR00827), and the VA VISN22 MIRECC. JRK is a founder and holds equity in Psynomics, Inc. The terms of this arrangement have been reviewed and approved by UCSD in accordance with its conflict of interest policies.




