Evidence for association between polymorphisms in the cannabinoid receptor 1 (CNR1) gene and cannabis dependence†
Please cite this article as follows: Agrawal A, Wetherill L, Dick DM, Xuei X, Hinrichs A, Hesselbrock V, Kramer J, Nurnberger JI Jr, Schuckit M, Bierut LJ, Edenberg HJ, Foroud T. 2009. Evidence for Association Between Polymorphisms in the Cannabinoid Receptor 1 (CNR1) Gene and Cannabis Dependence. Am J Med Genet Part B 150B:736–740.
Abstract
Genomic studies of cannabis use disorders have been limited. The cannabinoid receptor 1 gene (CNR1) on chromosome 6q14-15 is an excellent candidate gene for cannabis dependence due to the important role of the G-protein coupled receptor encoded by this gene in the rewarding effects of Δ9-tetrahydrocannabinol. Previous studies have found equivocal evidence for an association between SNPs in CNR1 and a general vulnerability to substance use disorders. We investigate the association between 9 SNPs spanning CNR1 and cannabis dependence in 1,923 individuals. Two SNPs that were previously associated with cannabis dependence in other studies were also significant with this phenotype in our analyses [rs806368 (P = 0.05) and rs806380 (P = 0.009)]. Haplotype analyses revealed the association to be largely driven by the SNP rs806380. These results suggest a role for the cannabinoid receptor 1 gene in cannabis dependence. © 2008 Wiley-Liss, Inc.
Between 1992 and 2002, cannabis use disorders increased by 18% in the United States [Compton et al., 2004]. The treatment episode data sets reveal that 289,532 (about 15%) of all admissions to chemical dependency treatment facilities in the United States were cannabis-related (Treatment Episode Data Set (TEDS), 2003). Additionally, cannabis use disorders are associated with the comorbid occurrence of alcohol dependence [Stinson et al., 2006], other drug dependence [Agrawal et al., 2007] and psychopathology [Grant, 1995; Compton et al., 2005; Copeland, 2006].
Family [Bierut et al., 1998; Merikangas et al., 1998] and twin studies [Kendler and Prescott, 1998; Tsuang et al., 2001; Agrawal and Lynskey, 2006] have demonstrated that genetic influences play an important role in shaping liability to cannabis dependence. Despite this, efforts to identify the genes that might contribute to this genetic liability have been limited. The cannabinoid receptor 1 gene (CNR1) on chromosome 6q14-15 encodes a G-protein coupled receptor for Δ9-tetrahydrocannabinol, and therefore is an excellent candidate gene for affecting susceptibility to cannabis dependence. The endogenous cannabinoid system is thought to regulate dopamine reward circuits that are believed to play an important role in reward processes involved in addiction [Mechoulam et al., 1994; Onaivi et al., 2002; Zhang et al., 2004]. The G-protein coupled receptor encoded by CNR1 is negatively coupled to adenylate cyclase, thus inhibiting cyclic AMP production. Further supporting the role of CNR1 in substance dependence is Rimonabant (SR141716A), a cannabinoid receptor 1 antagonist, that is being investigated for its potential role in alcohol, nicotine and drug dependence [Gelfand and Cannon, 2006; Cahill and Ussher, 2007; Benyamina et al., 2008; Jagerovic et al., 2008; Nides, 2008].
A recent linkage study identified the region on chromosome 6 in which this gene lies as linked with cannabis dependence [Agrawal et al., 2008b]. Previous studies have also reported evidence for the association between single nucleotide polymorphisms (SNPs) in CNR1 and a general vulnerability to substance use disorders. Zhang et al. 2004 tested for an association between 19 SNPs and the risk for polysubstance use disorders in European and African-American populations and found that three SNPs (rs806379-rs1535255-rs2023239) provided evidence of association when analyzed singly or as a haplotype (T-A-G haplotype). More recently, Hopfer et al. 2006 found that a SNP (rs806380—intron 2) proximal to the TAG haplotype was associated with the development of cannabis dependence symptoms (protective effect of G-allele) and that certain haplotypes (rs6454674-rs806380-rs806377-rs1049353) were associated with the development of fewer cannabis dependence problems while others were associated with an increased risk for cannabis dependence. More recently, SNPs in CNR1, especially rs6454674 and rs806368, were found to be associated with alcohol and illicit drug dependence in European Americans [Zuo et al., 2007]. Two recent studies also allude to the effects of SNPs in CNR1 on nicotine dependence and alcohol-related behaviors [Hutchison et al., 2008; Chen et al., 2008]. In contrast, the case control study of Herman et al. 2006 failed to find an association between cannabis, cocaine, opioid or polysubstance dependence and SNPs in CNR1. Negative association results have previously been reported by others [Li et al., 2000; Covault et al., 2001; Heller et al., 2001].
High levels of cannabis dependence have been reported in samples ascertained for alcohol dependence, including the Collaborative Study on the Genetics of Alcoholism (COGA) [Agrawal et al., 2008a]. Using a family based test of association, we tested the hypothesis that SNPs in CNR1 are associated with cannabis dependence in this sample.
Data collection occurred at six centers across the United States [Begleiter et al., 1995] and was approved by the institutional review boards of all participating centers. Probands were identified from alcohol treatment clinics. A subset of families with alcohol dependent probands as well as at least two first-degree relatives meeting criteria for both DSM-III-R alcohol dependence and Feighner criteria for definite alcoholism were selected for genotyping. We analyzed only the 219 families of self-reported European-American ancestry, which included 1,923 genotyped individuals. There were considerable differences between the European American and African American participants in the minor allele frequencies of SNPs in CNR1 (e.g., for rs806368, 20% in EA vs. 8% in AA; for rs1049353, 27% in EA vs. 7% in AA); therefore the African American families were excluded from all analyses.
Informed consent was obtained from all participants prior to the administration of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) interview [Bucholz et al., 1994; Hesselbrock et al., 1999]. DSM-III-R cannabis dependence [American Psychiatric Association, 1994] was coded dichotomously as “affected” if the participant endorsed 3 or more DSM-III-R dependence criteria (with the exclusion of withdrawal) and “unaffected” otherwise. Of the 1,923 genotyped participants (53% female), 379 met criteria for DSM-III-R cannabis dependence, 1,472 were unaffected (1,005 of which did not meet criteria for DSM-III-R alcohol dependence) and the remainder (N = 72) were missing data. The mean age of the cannabis dependent individuals in COGA was 33.1 [range 18–73] years while the unaffected individuals (neither alcohol nor cannabis dependence) had a mean age of 42.1 [range 18–92] years. Men were considerably more likely to meet criteria for cannabis dependence (30% vs. 12.4%, P < 0.0001) as were those with alcohol dependence (39% vs. 7.3%, P < 0.0001). Of those with cannabis dependence, 79.2% also met criteria for alcohol dependence. We also divided the sample into those aged 18–30 (lower 25%) and those aged 31 and older and computed the correlation between cannabis dependence and alcohol dependence. In the younger and older “cohorts,” the correlation between cannabis dependence and alcohol dependence was 0.61 and 0.65 respectively. This argues that there is no significant difference across age in the relationship between these disorders.
Eleven SNPs in CNR1 and in flanking regions were identified from publicly available databases (dbSNP, HapMap) and genotyped using a modified single nucleotide extensions reaction, with allele detection by mass spectometry (Sequenom MassArray system: Sequenom, San Diego, CA). All genotyped SNPs were checked for Mendelian inheritance using PEDCHECK [O'Connell and Weeks, 1998]. Marker allele frequencies and heterozygosities were computed using USERM13 [Boehnke, 1991]. One SNP, rs16880261, was monomorphic in this sample, and therefore was excluded from analyses. Tests for Hardy–Weinberg equilibrium, and linkage disequilibrium statistics, were computed in Haploview [Barrett et al., 2005]. SNP rs806377 had a significant deviation from Hardy–Weinberg equilibrium and was excluded from further analyses.
To test for the association between single SNPs and haplotypes with DSM-III-R cannabis dependence in the extended, multiplex COGA pedigrees, we used the Pedigree Disequilibrium Test (PDT) [Martin et al., 2001] implemented in pdtphase, part of the genetic analysis package UNPHASED [Dudbridge, 2003]. SNP physical positions were determined using NCBI build 36.1. For single SNP association analyses, both “sum” and “avg” (average) statistics were computed. The “sum” statistic allows for larger families to have greater weight than smaller families, while the “avg” statistic weighs all families equally in the computation of the test statistic. The “sum” statistic is more powerful when analyzing less prevalent disorders, such as cannabis dependence [Martin et al., 2001]. Hence, we only report single SNP and haplotype association results based on the “sum” statistics, which were largely similar to the statistical results obtained from the “avg” option. Haplotypes were constructed using sliding windows of varying width including 2–4 consecutive SNPs. Association analysis was carried out using UNPHASED. Rare haplotypes (frequency less than 5%) were excluded from the association analyses. In addition to utilizing the transmission of alleles from heterozygous parent to affected offspring, PDT statistics are also influenced by the transmission of alleles to the affected sibling in discordant sib-pairs.
Nine SNPs were analyzed across the 29 kb region of CNR1, extending 4 kb upstream of the gene (Fig. 1). D′ across adjacent SNPs was high (except between rs1049353 and rs2023239) while r2 ranged from 0.03 to 0.99. A similar pattern of LD was observed in the HapMap database. To determine whether the SNPs adequately assessed the variation within CNR1, the program Tagger [de Bakker et al., 2005] was used to examine the coverage by the nine SNPs. Seven of the nine were in the HapMap database. The average r2 of these seven SNPs with all 30 known HapMap SNPs (MAF ≥ 0.1) in the region was 0.66; r2 was greater than 0.5 for 80% of the SNPs and greater than 0.8 for 40% of the SNPs. Because two additional SNPs were genotyped, but were not in the HapMap database, our actual coverage of CNR1 was even better.
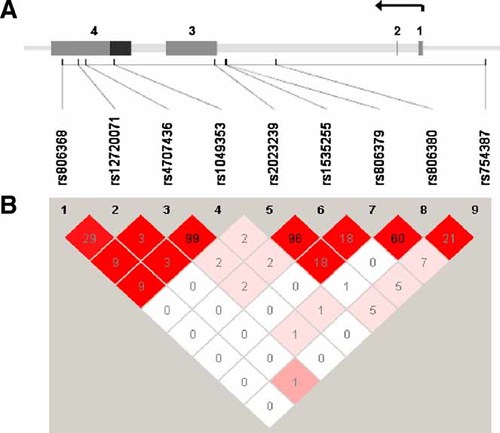
A: The gene structure of CNR1 was based on the annotation of Zhang et al. 2004, and is shown across the top of the figure with SNPs positioned below. Exons are numerically labeled with coding region in black and the untranslated regions in grey. The arrow denotes direction of transcription. B: Pairwise linkage disequilibrium between the genotyped SNPs is numerically represented as r2. The color coding reflects D′ with red reflecting a D′ = 1. The arrow represents the direction of transcription.
Results of the association analyses are provided in Table I. SNP rs806380 was significantly associated with cannabis dependence (P = 0.009) as was rs806368 (P = 0.05), while one SNP, rs806379, was marginally significant (P = 0.08). SNP rs806368 was in low to moderate LD with rs806379 and rs806380, suggesting independent effects. Because there might be some common genetic risk factors affecting several addictive disorders, we carried out secondary analyses in which we excluded (coded as unknown) from the sample of unaffected subjects those individuals who met criteria for DSM-III-R alcohol dependence. In these secondary analyses, the statistical significance was somewhat reduced (rs806380 P = 0.03) but the pattern of association was consistent.
| Marker name | Physical position (bp) | Allelesa | MAF | Z-statistic | PDT-sum P-value |
|---|---|---|---|---|---|
| rs806368 | 88,906,819 | C/T | 0.20 | 1.923 | 0.05 |
| rs12720071 | 88,907,900 | T/C | 0.08 | 0.759 | 0.45 |
| rs4707436 | 88,908,470 | A/G | 0.27 | 0.130 | 0.9 |
| rs1049353 | 88,910,354 | C/T | 0.27 | 0.130 | 0.9 |
| rs2023239 | 88,917,201 | C/T | 0.15 | 0.381 | 0.7 |
| rs1535255 | 88,917,927 | G/T | 0.15 | 0.591 | 0.55 |
| rs806379 | 88,917,986 | A/T | 0.47 | 1.770 | 0.08 |
| rs806380 | 88,921,372 | A/G | 0.35 | 2.608 | 0.009 |
| rs754387 | 88,935,578 | C/A | 0.28 | 1.136 | 0.26 |
- a The minor allele appears first (minor/major). SNPs in bold are statistically significant at P ≤ 0.05.
Two-SNP sliding window analyses found evidence for over-transmission of the G-C haplotype of rs806380 and rs754387 to unaffected individuals and over-transmission of the A-C haplotype to affected individuals (haplotype P-values 0.005, global P-value 0.004). Since the allele at rs754376 is the same in both haplotypes, we speculate that the association is primarily due to rs806380. No additional information was gained from increasing the window width. We also tested a haplotype formed by rs806368, rs806379, and rs806380. T-A-A was overtransmitted to affecteds (P = 0.06) while T-T-G was overtransmitted to unaffecteds (P = 0.06).
We demonstrate association between polymorphisms in the CNR1 gene (rs806380, rs806368, and rs754387) and DSM-III-R cannabis dependence. It should be noted that most of the cannabis dependent individuals in this sample were also alcohol dependent. Thus, the comorbid phenotype was the most common. There was no evidence for association of these SNPs with alcohol dependence per se. Our results suggest that variation in CNR1 may contribute to the risk of cannabis dependence. This finding is in keeping with previous studies of CNR1 and its influence on the liability to substance use disorders, however, these findings have been equivocal, with positive (P < 0.0001) [e.g., Zhang et al., 2004] and negative (P = 0.19–0.54) [e.g., Herman et al., 2006] results. In addition to consistency across sampling strategies [i.e., both Hopfer et al., 2006 and Zuo et al., 2007 used a case–control design], our results also replicate findings with specific SNPs. In an adolescent/young adult sample Hopfer et al. 2006 found that the G allele of rs806380 was overtransmitted to individuals (P = 0.034) with no symptoms of DSM-IV cannabis dependence (cases were defined as those with 1+ dependence symptoms). This fits well with our finding that the A allele was more common in cannabis-dependent individuals while the G allele was more common in those not meeting criteria for cannabis dependence. They also report that certain haplotypes (rs6454674, rs806380, rs806377, rs1049353: G-G-C-C) were associated with developing fewer dependence problems and others (T-A-C-C and G-A-C-C) were associated with an increased risk for cannabis dependence. Additionally, rs806368, marginally significant in our study, was found to significantly influence liability to substance dependence via an interaction with rs6454674 in an independent study [Zuo et al., 2007].
Although our data show that rs806380 provided the strongest evidence of association, that does not indicate it is a causative allele. There are seven known HapMap SNPs in CNR1 with a linkage disequilibrium estimate of r2 ≥ 0.50 with rs806380: six of which were not genotyped and one (rs806379, r2 = 0.60) which was marginally associated with cannabis dependence in our study.
Some limitations of the current study need to be considered. First, our analyses were restricted to the participants of European American ancestry—this association should be tested in other samples. Second, COGA ascertained densely affected multiplex families, and the genetic underpinnings of cannabis dependence in such a sample may not generalize to the population. Third, it was not possible to test for cannabis dependence exclusive of comorbid alcohol dependence, due to the high density of alcohol dependent individuals in COGA. However, even in a nationally representative survey of U.S. adults (NESARC) [Grant et al., 2003], 69.3% of those with cannabis dependence also met criteria for alcohol dependence, suggesting similar levels of comorbidity in the general population. A fourth limitation is that rs806380, with which we detect significant association, had a Hardy–Weinberg P-value of 0.01; however, there was no evidence for an increased dropout rate (1.6%) and the observed heterozygosity was 0.46, close to the predicted value. Replication studies will, as always, prove invaluable.
In summary, this sample ascertained for alcoholism validates converging evidence from several independent studies suggesting that polymorphisms in CNR1 may be associated with cannabis dependence. The action of these polymorphisms, which so far have not been attributed to any structural or functional effects on cannabis use and misuse, is still unknown. Our findings, in addition to the extant literature, encourage further research into the etiologic contributions of the CNR1 gene to cannabis dependence.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Co-Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., P.M. Conneally, T. Foroud); University of Iowa (S. Kuperman, R. Crowe); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Zhaoxia Ren serves as the NIAAA Staff Collaborator. This national collaborative study is supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). Arpana Agrawal is also supported by DA023668. In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal Investigators of COGA since its inception; we are indebted to their leadership in the establishment and nurturing of COGA, and acknowledge with great admiration their seminal scientific contributions to the field.




