Association analysis of NOTCH 4 polymorphisms with schizophrenia among two independent family based samples
Abstract
The present study investigated polymorphisms of the NOTCH 4 gene in two independent samples from India and USA, consisting of patients with schizophrenia and their parents (n = 182, and n = 148 ‘trios,’ respectively). Five DNA markers, namely (GAAG)n, (TAA)n, SNP1, SNP2, and (CTG)n were evaluated. Transmission distortion, consistent with a modest association was detected among both samples. Additional association studies at this locus are warranted. © 2004 Wiley-Liss, Inc.
INTRODUCTION
The etiology of schizophrenia is multi-factorial, with the possibility of several susceptibility genes conferring modest risk. Despite promising results, it has been difficult to identify genes that confer risks of a relatively small magnitude for common human diseases. Typically, promising associations have not been replicated in follow-up studies having sufficient power [Dahlman et al., 2002]. The initial reports tend to overestimate the effect size and artifacts can undoubtedly inflate the initial results [Cohen, 1999]. On the other hand, considerable inter-study heterogeneity contributes to difficulties with replications. Thus, meta-analyses may be required to resolve inconsistencies [Ioannidis et al., 2001]. Unfortunately, reluctance to publish non-significant associations may lead to premature loss of interest in a ‘true’ susceptibility gene. We illustrate this problem with respect to a controversial association between NOTCH 4 and schizophrenia.
Linkage and association studies have been instrumental in the effort to map schizophrenia susceptibility genes. In particular, genome wide linkage scans have repeatedly identified 6p21-24 region as a putative susceptibility region [Antonarakis et al., 1995; Schwab et al., 1995; Straub et al., 1995; Wang et al., 1995; Turecki et al., 1997; Lindholm et al., 1999; Bailer et al., 2000; Schwab et al., 2000; Lewis et al., 2003]. A detailed account of studies in this region has been published recently by Skol et al. [2003]. The reported association of schizophrenia with NOTCH 4 [Wei and Hemmings, 2000] and Dysbindin (DTNBP1) genes [Straub et al., 2002], localized in this region are consistent with the linkage results. NOTCH family of genes are involved in inter-cellular signaling and encode trans-membrane receptors. Some human diseases have been found to be associated with NOTCH mutations; for example, leukemia and CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) [Joutel and Tournier-Lasserve, 1998] [reviewed by Kageyama and Ohtsuka, 1999].
Wei and Hemmings [2000] reported a significant association of schizophrenia with polymorphism at NOTCH 4 gene, located within the HLA class III region. The Transmission Disequilibrium Test (TDT), which detects association only in the presence of linkage, revealed significant transmission distortion for three NOTCH 4 markers in this study. A subsequent large international consortium detected a trend for over-transmission of 13 repeat allele of (TAA)n marker and 11 repeat allele of (CTG)n to schizophrenia patients in German-Israeli sib pair samples [Sklar et al., 2001]. Investigation of these markers in a large Chinese sample did not reveal any association [Fan et al., 2002]. In addition, four other case-control studies failed to detect significant association [Imai et al., 2001; McGinnis et al., 2001; Ujike et al., 2001; Wassink et al., 2003]. Thus, a resounding argument for absence of a genetic association could be made. Nonetheless, each of these studies evaluated only a fraction of all the identified polymorphisms at the NOTCH 4 gene, which spans 56.8 kb. Given the highly variable and uncertain linkage disequilibrium observed over small distances in the human genome, identification of the primary association conferring susceptibility could arguably be elusive [Martin et al., 2000]. Plausible functional correlations have recently been proposed for the putative risk allele(s). While it has long been known that NOTCH 4 is expressed in the human brain [Uyttendaele et al., 1996], allelic associations of a (CTG)n polymorphism with brain volumes estimated from MRI scans, as well as performance on the Wisconsin Card Sort Test (WCST) were recently reported among individuals with schizophrenia, as well as unaffected controls [Wassink et al., 2003]. These analyses suggest credible mechanisms of pathogenesis. With this background, we investigated associations at NOTCH 4 in two large and independent family based samples.
MATERIALS AND METHODS
Clinical Analysis
Participants were recruited through ongoing genetic epidemiological studies at Pittsburgh and New Delhi, using identical assessment procedures. The sample includes 182 case-parent trios from India and 148 trios from USA (total 330 trios). The probands were evaluated using the English or Hindi versions of the Diagnostic Interview for Genetic Studies (DIGS). In conjunction with information from medical records and information from relevant relatives, consensus diagnoses were established using DSM IV criteria [please see Chowdari et al., 2001, for details]. All participants provided written informed consent in accordance with guidelines from the University of Pittsburgh Institutional Review Board, as well as the Ethics Committees at Delhi University and at Dr. Ram Manohar Lohia Hospital, New Delhi.
Laboratory Analysis
Five polymorphisms were analyzed using PCR based assays. The list included four from the initial report [two single nucleotide polymorphisms: SNP1 and SNP2; two trinucleotide repeat markers (CTG)n and (TAA)n] [Wei and Hemmings, 2000]. An additional (GAAG)n tetranucleotide repeat [reported by Li et al., 1998] that was not analyzed in this study was also included. Our BLAST searches of available genomic sequence databases revealed that the physical order of this marker is different from that reported earlier by Li et al. [1998] (please see Fig. 1 for current alignment). Further, the SNP1 and SNP2 markers reported by Skol et al. [2003] seem to be different from that analyzed originally by Wei and Hemmings [2000] and also analyzed in our study (base pair positions marked in Fig. 1). Genomic DNA was isolated from 10 ml venous blood using the phenol chloroform extraction method at the respective laboratories in New Delhi (BKT) and Pittsburgh (VLN). Details of the primers, PCR conditions as well as the method of analysis for each of the analyzed markers are available on the web (http://www3.interscience.wiley.com/cgi-bin/jhome/99018626). The (TAA)n marker was not analyzed in the Indian samples.
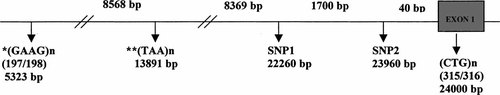
NOTCH 4 polymorphisms. *, This marker is named GGAA elsewhere and its localization differs from prior publications. Its placement here is based on a BLAST of clone U89335 and the consensus MHC sequence using the specified primers. **, Not analyzed in the Indian sample.
Statistical Analysis
Genotypes for all the markers were tested for Hardy–Weinberg equilibrium and Mendelian inconsistencies. The Genehunter program has been used to perform TDT analysis using individual SNPs and related haplotypes [Kruglyak et al., 1996]. MCETDT for single locus TDT for the polymorphic markers was also used [Zhao et al., 1999].
RESULTS
All genotypes were in Hardy–Weinberg equilibrium, apart from those involving SNP1 among the US cases (P = 0.0003) and the (CTG)n repeat in the Indian sample (P < 0.00001). TDT analysis of individual polymorphisms suggested modest associations for certain alleles of the (CTG)n and the (GAAG)n STRPs, though permutation tests (implemented in Genehunter) incorporating all alleles and a separate MCETDT analysis for single locus TDT did not suggest overall transmission distortion. TDT results for markers with nominal transmission distortion (P < 0.05, uncorrected) are presented in Table I.
| Marker | Allele (repeats) | India samples (n = 182 trios) | USA samples (n = 148 trios) | India + USA samples (n = 330 trios) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | U | χ2 | P value | T | U | χ2 | P value | T | U | χ2 | P value | ||
| (CTG)n | 2 (6) | 8 | 20 | 5.14 | 0.023 | 13 | 14 | 0.04 | 0.847 | 21 | 34 | 3.07 | 0.080 |
| 6 (10) | 51 | 38 | 1.90 | 0.168 | 38 | 23 | 3.69 | 0.055 | 89 | 61 | 5.28 | 0.022 | |
| (TAA)n* | 3 (8) | — | — | — | — | 20 | 32 | 2.77 | 0.09 | — | — | — | — |
| 8 (13) | — | — | — | — | 4 | 12 | 4.0 | 0.046 | — | — | — | — | |
| 197/198 (GAAG)n | 10 (46) | 5 | 6 | 0.09 | 0.763 | 4 | 12 | 4.0 | 0.046 | 9 | 17 | 2.46 | 0.117 |
| 14 (50) | 13 | 5 | 3.56 | 0.059 | 7 | 2 | 2.78 | 0.09 | 20 | 7 | 6.26 | 0.012 | |
| 18 (54) | 6 | 0 | 6 | 0.014 | 8 | 6 | 0.29 | 0.593 | 14 | 6 | 3.20 | 0.074 | |
- T, transmitted; U, untransmitted; *, not analyzed in Indian sample.
- No association with SNP1 and SNP2 alleles and hence not included.
Haplotype based analyses were also conducted. Though significant transmission distortion was not observed at any of the markers following corrections for multiple comparisons (mentioned above), nominally significantly increased or reduced transmissions were observed when individual haplotypes were compared with all the other haplotypes. Such results (P < 0.05, uncorrected) are presented in Table II. By and large, transmission distortion was observed at haplotypes consisting of several combinations of markers, usually with differing allelic combinations in the Indian and the US samples. The (CTG)n marker and the adjacent SNP2 revealed transmission distortion in the Indian, as well as US samples, albeit with different haplotypes. The haplotype bearing the ten repeat (allele 6) of (CTG)n and allele 2 of SNP2 was over-transmitted in the Indian sample (transmitted: 39, not transmitted: 22, χ2 = 4.74, 1 df, P = 0.029) and a trend was also present in the US sample (transmitted: 27, not transmitted: 17, χ2 = 2.27, 1 df, P = 0.13). Excess transmission of this haplotype was also observed in the combined sample (transmitted: 66, not transmitted: 39, χ2 = 6.94, 1 df, P = 0.008).
| Haplotype | Allelea | India samples (n = 182 families) | USA samples (n = 148 families) | India + USA samples (n = 330 families) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t | U | χ2 | P value | t | U | χ2 | P value | t | U | χ2 | P value | ||
| SNP-1 and SNP-2 | 1,1 | 8 | 20 | 5.14 | 0.023 | ||||||||
| SNP-2 and (CTG)n | 2,6 | 39 | 22 | 4.74 | 0.029 | 27 | 17 | 2.27 | 0.132 | 66 | 39 | 6.94 | 0.008 |
| 2,9 | 0 | 4 | 4 | 0.046 | |||||||||
| 1,6 | 0 | 5 | 5 | 0.025 | 0 | 1 | 1 | 0.317 | 0 | 6 | 6 | 0.014 | |
| SNP-1, SNP-2, and (CTG)n | 1,2,6 | 19 | 15 | 0.47 | 0.493 | 27 | 13 | 4.90 | 0.027 | 46 | 28 | 4.38 | 0.036 |
| 2,2,5 | 1 | 8 | 5.44 | 0.020 | |||||||||
| 1,2,9 | 0 | 4 | 4 | 0.046 | |||||||||
| 197/198, SNP-1 | 10,1 | 1 | 4 | 1.80 | 0.180 | 0 | 5 | 5 | 0.025 | 1 | 9 | 6.40 | 0.011 |
| 15,1 | 0 | 1 | 1 | 0.317 | 1 | 6 | 3.57 | 0.059 | 1 | 7 | 4.50 | 0.034 | |
| 17,1 | 0 | 1 | 1 | 0.317 | 0 | 4 | 4 | 0.046 | 0 | 5 | 5.0 | 0.025 | |
| 25,1 | 2 | 7 | 2.78 | 0.096 | 0 | 4 | 4 | 0.045 | 2 | 11 | 6.23 | 0.013 | |
| 197/198, SNP-1, and SNP-2 | 10,1,2 | 1 | 3 | 1.00 | 0.317 | 0 | 4 | 4 | 0.046 | 1 | 7 | 4.50 | 0.034 |
| 15,1,1 | 0 | 5 | 5 | 0.025 | |||||||||
| 25,1,2 | 1 | 4 | 1.8 | 0.180 | 0 | 4 | 4 | 0.046 | 1 | 8 | 5.44 | 0.02 | |
| (TAA)n, SNP-1 | 3,1 | 5 | 15 | 5 | 0.025 | ||||||||
| 8,1 | 0 | 7 | 7 | 0.008 | |||||||||
| (TAA)n, SNP-1, and SNP-2 | 3,1,2 | 0 | 4 | 4 | 0.046 | ||||||||
| 4,1,1 | 0 | 4 | 4 | 0.046 | |||||||||
| 197/198 and (TAA)n | 15,3 | 0 | 4 | 4 | 0.046 | ||||||||
| 15,5 | 4 | 0 | 4 | 0.046 | |||||||||
- a Allele code.
DISCUSSION
The present study investigated genetic association in the NOTCH 4 gene region using two independent samples from US and India. We investigated five polymorphisms, four of which were described in the original study by Wei and Hemmings [2000]. Our analyses do not conclusively prove an association. Nevertheless, it is difficult to ignore several suggestive trends.
Our analyses suggest associations at nominal levels of significance, as well as suggestive trends for associations with several individual polymorphisms in both the Indian and the US samples (Table I). None of these results remained significant following corrections for multiple comparisons, but are consistent with the results from the initial report [Wei and Hemmings, 2000]. We observed suggestive associations of the (CTG)n polymorphism, albeit with different alleles in the two samples (six repeat allele among Indians and ten repeat allele among US samples, Table I). In addition, there was significant association with allele 14 of (GAAG)n in the pooled sample (P = 0.012). Besides, analysis of the 54 repeat allele of (GAAG)n also showed a significant association with the Indian samples (P = 0.01) and a suggestive trend is present in the combined data set (P = 0.07). Similarly, though allele 2 of SNP2 alone does not show any independent allelic association, it is present in the associated haplotypes in both the India and the US samples along with (CTG)n allele. In contrast to the studies by Sklar et al. [2001] and Skol et al. [2003], we did not find any significant association with any of the (TAA)n alleles in the US population.
We also observed trends for transmission distortion of haplotypes incorporating several sets of markers. The most notable over-transmission was observed using a haplotype at SNP2 and (CTG)n (alleles 2 and 6, respectively) among the Indian trios (P = 0.029), and in the pooled sample (P = 0.008), with a suggestive trend in the US sample (P = 0.132). It is notable that excess transmission of haplotypes based on SNP2/(CTG)n polymorphisms was also noted in the initial report from Caucasian samples in the UK [Wei and Hemmings, 2000]. Consistent with these results, a haplotype encompassing SNP1-SNP2-(CTG)n (alleles 1, 2, and 6, respectively) was over-transmitted in the US sample (P = 0.027) and in the pooled samples (P = 0.036). Though significant associations are seen with some of the other haplotypes as detailed in Table II, caution is necessary, in view of the small number of transmissions observed in each of these analyses. However, it is noteworthy, that neither the same allele nor the same haplotype is significantly associated in Indian and US samples and in the combined samples, indicating heterogeneity between the two populations.
Recently, excess transmission of eight and 13 repeat alleles of the (TAA)n marker was reported among African-American patients with schizophrenia [Skol et al., 2003]. Separately, the 11 repeat allele of (CTG)n was shown to be associated with schizophrenia among German-Israeli sibpairs [Sklar et al., 2001]. Though no association of (CTG)n with schizophrenia was reported among Japanese [Imai et al., 2001], a recent family based study on Japanese and Chinese samples suggest a possible association of Notch 4 with early onset schizophrenia or schizophrenia with negative symptoms [Takahashi et al., 2003]. All these observations suggest that association of Notch 4 gene markers (such as SNP2, (CTG)n, and (TAA)n) with schizophrenia may be present but with different alleles and to varying extents in different populations.
Thus, our analyses, in conjunction with all the other published reports suggest that it may be premature to discard NOTCH 4 gene as a susceptibility gene for schizophrenia. Given the variable and uncertain linkage disequilibrium observed over small distances in the human genome, more polymorphisms may need to be evaluated at this locus. If present, associations will almost certainly be modest. Meta-analysis may thus be essential to resolve the putative association, and publication of results from such samples is necessary.




