Distal 1q Duplication and Distal 9p Deletion: A Follow-Up Case Report and Literature Review on Candidate Genes for 9p Deletion Syndrome
Funding: The authors received no specific funding for this work.
ABSTRACT
Distal 1q duplication and distal 9p deletion are rare chromosomal aberrations associated with developmental delay and mild to moderate congenital malformations. There are inconsistent findings regarding the critical region for trigonocephaly within 9p deletion syndrome. A recent analysis of the largest 9p- cohort to date, however, delineated two critical regions and emphasized the need for replication. We report on a trigonocephalic child with a de novo 46.09 megabases (Mb) terminal duplication of 1q and a 5.31 Mb terminal deletion in 9p, described as 46,XX,der(9)t(1;9)(q32.1;p24.1). The clinical course was predominantly influenced by the 1q duplication. Trigonocephaly, however, was consistent with 9p deletion syndrome. Our findings support the delineation of [GRCh38] 9:3,418,241–5,341,746 as a critical region for trigonocephaly within 9p deletion syndrome. We propose that haploinsufficiency of RFX3, along with complex gene interactions, contributes to the mechanism for disease.
1 Introduction
Congenital genetic malformations affect approximately 3% of live births. With chromosomal aberrations accounting for an estimated 75% of cases with known etiology (Feldkamp et al. 2017), first-tier genetic testing includes Giemsa-banded karyotyping (GTG), fluorescence in situ hybridization (FISH), and chromosomal microarray analysis (CMA) (Miller et al. 2010). Using these methods, both classic aneuploidies (e.g., trisomy 21) and rarer segmental aneuploidies (e.g., distal 1q duplication [1q+] and distal 9p deletion [9p−]) resulting from unbalanced chromosomal rearrangements can be detected. Similar to other copy number variations, segmental aneuploidies are frequently associated with developmental delay (DD), poor growth, and malformations of varying specificity.
Selected malformations associated with 1q+ and 9p− are summarized in Table 1. Regarding 1q+, screening and management of respiratory insufficiency and dysphagia are crucial, particularly during infancy (Nowaczyk et al. 2003). Respiratory symptoms tend to improve with age (Watanabe et al. 2016), and survival into adulthood has been reported (Van Buggenhout et al. 1998). The adult phenotype comprises intellectual disability (ID) with a need for assistance in daily activities, along with basic verbal and more elaborate nonverbal communication skills (Meloni et al. 2012).
| 1q+ a | 9p− b | Present case | |
|---|---|---|---|
| Developmental delay | + | + | + |
| Hypotonia | − | + | + |
| Brain malformation | + | +/− | + c |
| Heart defect | + | + | + d |
| Recurrent respiratory tract infection | + | + | + |
| Differences in sex development | − | + | − |
| Dysphagia | + | − | + |
| Nystagmus | − | +/− | + |
| Strabismus | − | +/− | + |
| Short stature | +/− | − | + |
| Dysmorphic features | |||
| Skull | |||
| Trigonocephaly | − | + | + |
| Midface retrusion | +/− | + | + |
| Prominent forehead | + | − | + |
| Eyes | |||
| Highly arched eyebrows | − | + | − |
| Epicanthus | − | + | − |
| Downslanted palpebral fissures | + | − | + |
| Closely spaced eyes | − | − | + |
| Blepharophimosis | − | +/− | + |
| Nose, mouth, and ears | |||
| Depressed nasal bridge | + | + | + |
| Long philtrum | + | + | + |
| Thin upper lip | +/− | + | + |
| High palate | +/− | + | + |
| Narrow mouth | +/− | + | + |
| Low-set ears with increased posterior angulation | + | + | + |
| Thorax, neck, and extremities | |||
| Wide intermamillary distance | +/− | + | + |
| Short neck | +/− | + | + |
| Tapered fingers | +/− | + | + |
| Camptodactyly | +/− | +/− | + |
| Overlapping toes | +/− | +/− | + |
- Abbreviations: −, rarely present or absent; +, (frequently) present; +/−, infrequently present; 1q+, distal 1q duplication; 9p−, distal 9p deletion.
- a Watanabe et al. (2016) and Nowaczyk et al. (2003).
- b Starosta et al. (2024) and Sams et al. (2022).
- c Corpus callosum hypoplasia and pachygyria.
- d Atrial septal aneurysm with reduced left ventricular ejection fraction.
With regard to 9p−, terminal deletion 9p24.3 (MIM: 154230) is distinguished from larger deletions in 9p (9p deletion syndrome, MIM: 158170). Terminal deletion 9p24.3 is primarily associated with gonadal dysgenesis, particularly in 46,XY individuals. 9p deletion syndrome, by contrast, is linked to DD and distinct malformations, including trigonocephaly (Starosta et al. 2024). Individuals with 9p− often also require support for daily activities but may attend special education programs and acquire basic reading and writing skills (Swinkels et al. 2008). First described by Alfi et al. 1973, numerous studies have sought to delineate the critical region and propose candidate genes (Ajami et al. 2023; Alfi et al. 1973; Christ et al. 1999; Hauge et al. 2008). A recent analysis of the largest 9p− cohort to date, utilizing genome sequencing, identified two critical regions for trigonocephaly: a primary region at [GRCh38] 9:14,550,892–17,102,299 (including FREM1) and a secondary region at [GRCh38] 9:3,418,241–5,341,746 (Starosta et al. 2024).
In this report, we focus on 9p deletion syndrome, comparing clinical and molecular (cyto-) genetic findings from our case with those from previously reported cases. Our objective is to further refine the critical region for trigonocephaly and to propose candidate genes.
2 Material and Methods
Karyotyping was performed by Giemsa-trypsin banding on peripheral blood leukocytes, following standard procedures. For CMA, genomic DNA was isolated from peripheral blood leukocytes using the QIAamp DNA Mini Blood Kit (Qiagen GmbH, Hilden, Germany). The CytoSure Constitutional V3 platform (OGT, Oxford, UK), containing approximately 180,000 genome-wide oligonucleotides, was used according to the manufacturer's protocol. The array slide was scanned with an Innoscan 910 scanner, and data extraction was carried out using Mapix software (Innopsys, Carbonne, France). The resulting data were analyzed using CytoSure Interpret Software (version 4.11.30, OGT, Oxford, UK). Clinically relevant mosaicism was excluded through FISH on buccal mucosal cells, using subtelomere-specific DNA probes for chromosomes 1qter (GDB:315525; Cytocell, Sysmex Europe SE) and 9pter (RH65569, Cytocell, Sysmex Europe SE).
A literature review on the critical region for 9p deletion syndrome and associated candidate genes was conducted using MEDLINE, Embase, and Web of Science. Included were studies on patients with 9p− who presented with trigonocephaly, as detected by molecular (cyto-)genetic testing. Detailed inclusion and exclusion criteria, along with a flow diagram, are provided in SI S1. ClinGen haploinsufficiency score (little, some, or sufficient evidence) and gnomAD loss of function observed/expected upper bound score (< 0.6) were used to assess likely haploinsufficient genes, with at least one criterion to be met.
3 Case Presentation
The patient was first evaluated during the neonatal period and underwent repeated assessments over the following 2 years. Following gestational diabetes, delivery was by cesarean section at 33 + 2 weeks gestational age due to premature rupture of membranes (APGAR 9/10/10). Birth weight was 1460 g (7th centile, small for gestational age), length was 41 cm (13th centile), and head circumference was 30 cm (29th centile). Dysmorphic features and primary clinical findings are detailed in Table 1. Laboratory tests indicated central hypothyroidism, growth hormone deficiency, and elevated gamma-glutamyl transferase. For patient image, see SI S2.
Due to feeding difficulties, nutrition had to be provided via a nasogastric tube. Beginning on day four, the patient experienced recurrent episodes of oxygen desaturation accompanied by bradycardia. Management included high-flow nasal cannula oxygen therapy and administration of caffeine for apnea of prematurity. She was discharged from the neonatal intensive care unit on day 60 in stable condition with ongoing oxygen therapy and nasogastric feeding. The patient was readmitted at 6, 9, 11, 12, and 13 months of age due to pneumonia. Long-term respiratory management included nocturnal oxygen therapy at 9 months and subsequently, nocturnal noninvasive ventilation without supplemental oxygen at 15 months. By 13 months of age, feeding difficulties, including severe dyschezia, had become a major concern. Fiberoptic endoscopic evaluation of swallowing identified mild to moderate dysphagia, leading to the placement of a percutaneous endoscopic gastrostomy (PEG) tube, while small amounts of oral nutrition were maintained. At 2 years of age, growth parameters showed a weight of 10.41 kg (16th centile, corrected age), length of 80 cm (4th centile), and a head circumference of 47.5 cm (30th centile). Cognitive-motor skills included vocalizing, social smiling, rolling over, grasping objects, and brief eye contact. However, she did not achieve head control and remained unable to crawl, sit independently, walk, play, or speak. Overall, findings indicated profound DD, with ongoing requirements for PEG tube feeding and nocturnal noninvasive ventilation.
GTG revealed an unbalanced karyotype with a derivative chromosome 9. CMA showed a terminal 46.09 Mb duplication of 1q32.1q44 and a terminal 5.51 Mb deletion in 9p24.1p24.3, resulting in the karyotype arr[hg19] 1q32.1q44(203125049_249212666)x3,9p24.3p24.1(204090_5517253)x1 (see SI S3). Likely haploinsufficient genes in 9p were CD274, DMRT1, RFX3, and SMARCA2 (for a list of all deleted protein-coding OMIM genes, see SI S4). Parental GTG showed normal results (data not shown), indicating that the unbalanced translocation likely arose de novo with a small possibility of parental germline mosaicism.
4 Discussion
To our knowledge, this is the first reported case of a patient with a 46,XX,der(9)t(1;9)(q32.1;p24.1) karyotype. Compared to previous reports on unbalanced rearrangements involving 1q and 9p (with breakpoints at 1q41, 1q42, and 1q44, respectively), the 1q duplication in our case is relatively large (Abreu et al. 2014; Vahabi et al. 2017; Verbraak et al. 1992). The deletion in 9p, however, is among the smallest terminal deletions associated with trigonocephaly reported to date (Figure 1).
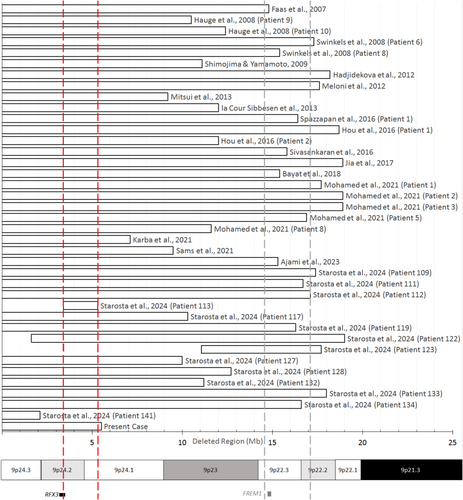
Bars represent deleted regions in megabases (Mb) as defined by molecular (cyto-)genetic testing. The dashed vertical lines indicate the primary (gray) and secondary (red) critical regions for trigonocephaly, as defined by Starosta et al. (2024). The ideogram illustrates bands of 9p, FREM1 as the most prominent candidate gene within the primary critical region, and RFX3 as the only likely haploinsufficient gene within the secondary critical region.
Our findings underline the phenotypic predominance of distal 1q duplications in complex segmental aneuploidies (Barros-Núñez et al. 1989). Respiratory insufficiency and dysphagia were the predominant symptoms, necessitating long-term PEG tube feeding and nocturnal noninvasive ventilation. A reduced hospitalization rate and the acquisition of basic non-verbal communication skills over time were consistent with other reports describing gradual improvement in respiratory function and cognitive-motor development in patients with distal 1q duplication (Watanabe et al. 2016).
Particularly, trigonocephaly, however, was consistent with 9p deletion syndrome. Historically, the proposed critical region and associated candidate genes have shown significant variability across studies. Potential explanations for these discrepancies include varying clinical definitions (e.g., differences in sex development, ID, dysmorphic features), methodological variations in genetic testing techniques (e.g., GTG, FISH, CMA, exome sequencing, genome sequencing), and the contribution of complex gene interactions. To enhance comparability, we limited our review to patients presenting trigonocephaly as the most distinct feature of 9p deletion syndrome. Additionally, to reduce methodological bias, we included molecular (cyto-)genetic studies only.
The 5.51 Mb deleted region in the present case encompasses frequently proposed candidate genes for DD (e.g., SMARCA2) and gonadal dysgenesis (DMRT1, DMRT3) (Ajami et al. 2023; Sams et al. 2022). In contrast, FREM1, the most prominent candidate gene for trigonocephaly, is located outside the deleted region (Ajami et al. 2023; Swinkels et al. 2008; Vissers et al. 2011). Our findings, supported by a comprehensive literature review, align with the delineation of [GRCh38] 9:3,418,241–5,341,746 as a critical region for trigonocephaly, as defined by Starosta et al. (2024) (Figure 1). Subsequently, we screened for haploinsufficient genes within this region that could contribute to trigonocephaly (SI S4). Aside from 9p deletion syndrome, genetic alterations associated with syndromic trigonocephaly include various chromosomal aberrations (primarily deletions such as deletion 11q in Jacobsen syndrome) as well as pathogenic monogenic variants (e.g., CD96 in C syndrome) (Goos and Mathijssen 2019). The common underlying mechanism may involve disrupted signaling pathways, such as fibroblast growth factor (FGF)/FGF receptor (FGFR) and Hedgehog signaling, resulting in altered stem cell migration and impaired osteogenic differentiation (Wu and Gu 2019). Among the likely haploinsufficient genes, RFX3 in particular may therefore contribute to metopic synostosis by regulating ciliogenesis: RFX3 encodes a transcription factor essential for the formation and function of primary cilia. Cilia, in turn, play a pivotal role in mediating and regulating FGF/FGFR and Hedgehog signaling, both of which are critical for cranial suture development (Sugiaman-Trapman et al. 2018; Tabler et al. 2013). It should be noted, however, that loss-of-function variants in RFX3 have been associated with nonspecific signs commonly observed in 9p deletion syndrome (e.g., hypotonia, high palate, tapered fingers), but not with trigonocephaly (Harris et al. 2021).
In general, pathogenicity models in deletion syndromes involve haploinsufficiency, either of a single causative gene (e.g., RAI1 in Smith-Magenis syndrome) or of multiple adjacent genes (contiguous gene syndromes, e.g., 7q11.23 deletion in Williams syndrome), as well as more complex gene interactions such as epistasis (e.g., 16p11.2 deletion) (Iyer et al. 2018). The absence of a clearly defined critical region, despite efforts to mitigate bias from methodological limitations and varying clinical definitions, suggests that complex gene interactions most likely contribute to the mechanism for disease in 9p deletion syndrome. In support of this hypothesis, previous studies have demonstrated a significant enrichment of protein–protein interactions among 9p genes, suggesting a complex genetic network (Sams et al. 2022).
Major limitations of this non-systematic review include the absence of high-resolution genomic data (e.g., genome sequencing) and functional studies.
In summary, our findings support the delineation of [GRCh38] 9:3,418,241–5,341,746 as a critical region for trigonocephaly within 9p deletion syndrome, as recently proposed. Aside from FREM1, haploinsufficiency of RFX3, along with complex gene interactions, may contribute to the mechanism of disease. We suggest follow-up studies focusing on functional analyses of RFX3 in particular, including transcriptome profiling.
Author Contributions
All authors have accepted responsibility for the entire content of this manuscript and approved its submission. The authors applied the SDC approach for the sequence of authors. Jonas Helbig: conceptualization, writing – original draft, and writing – review and editing. Jürgen Kunz: methodology. Anca Mannhardt, Ellen Gandaputra: writing – review and editing. Christiane Kling: supervision and writing – review and editing.
Ethics Statement
This study was performed in accordance with the Declaration of Helsinki principles. IRB approval was not required for this single case report. Written informed consent was obtained from the patient's parents for publication of this case report and any accompanying images.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




