Expanded phenotypic spectrum of UDP-glucose-6-dehydrogenase recessive neurodevelopmental disorder: Two novel descriptions with or without epileptic encephalopathy
[Correction added after first online publication on 2 August 2024. Author name “Pauline Bordeneuve-Plante” has been changed to “Pauline Plante-Bordeneuve”.]
Abstract
Recent advances in the understanding of infantile developmental epileptic encephalopathies (IDEE) have revealed the association of biallelic pathogenic variants in UGDH. In this study, we report two novel combinations identified by exome sequencing: p.(Arg135Trp) with p.(Arg65*) and p.(Arg102Trp) with p.(Arg65*). Both combinations share a common pathogenic nonsense variant, with the missense variants strategically located in the NAD-binding domain of the UGDH protein, predicted in structural models to create new interactions with the central domain. The first patient exhibited the typical UGDH-related disease phenotype and progressive microcephaly, a rarely reported feature. In contrast, the second patient presented an atypical phenotype, including absence of seizure, severe intellectual disability, ataxic gait, and abnormal eye movements. This comprehensive analysis extends the phenotypic spectrum of UGDH syndrome beyond early infantile intractable encephalopathy to include intellectual disability without epilepsy.
1 INTRODUCTION
The UGDH gene (MIM#603370) encodes the UDP-glucose dehydrogenase, a critical enzyme in the glycosaminoglycan (GAG) biosynthesis pathway, responsible for the synthesis of UDP-glucuronic acid (UDP-GlcA) from UDP-glucose (UDP-Glc) (Egger et al., 2011). The UGDH enzyme is composed of three domains, two structurally homologous α/β structures, the N-terminal/NAD-binding and the C-terminal/UDP-binding, and an α-helical central domain (Rajakannan et al., 2011). The structure is assembled in a hexameric disk shaped, with a trimer of dimers required for enzymatic function. The conversion of UDP-Glc to UDP-GlcA by UGDH involves the concomitant reduction of NAD+ to NADH, a critical step in the process of nucleotide sugar interconversion. UDP-GlcA serves as a precursor for the synthesis of GAGs, such as chondroitin sulfate, dermatan sulfate, keratan sulfate, and heparan sulfate, which are major components of the proteoglycans in the extracellular matrix (Clarkin et al., 2011). UDP-GlcA is also involved in the detoxification process through glucuronidation, where it conjugates with various endogenous and exogenous compounds for excretion (Egger et al., 2011).
Glycosylation defects have been observed in more than 100 rare human genetic disorders, affecting the central and/or the peripheral nervous system. Common clinical manifestations include developmental delay or intellectual disability, seizures, neuropathy, and metabolic abnormalities in multiple organ systems (Mizumoto & Yamada, 2021). Biallelic loss-of-function pathogenic variants in the UGDH gene have been described in patients with recessive infantile developmental epileptic encephalopathies (IDEE or Jamuar syndrome; MIM#618792). Thus, genetic defects in the UGDH gene have been associated with marked developmental delay, epilepsy, mild nonspecific dysmorphism, and motor disorder with axial hypotonia. Seizures were observed in all reported patients. The first reported patient even presented a pharmacoresistant epilepsy (Hengel et al., 2020).
Here, we report on the identification of compound heterozygous variants in the UGDH gene in two patients, one of whom had no history of seizures.
2 METHODS
2.1 Exome sequencing and data processing
Blood samples were collected from the affected individuals and their parents after informed consents. Samples were prepared using Illumina DNA Prep Enrichment (Illumina, San Diego, US-CA). A 150 bp paired-end exome sequencing was performed on an Illumina NovaSeq 6000 platform, with an average on-target coverage of 150× and analyzed using GATK best practices. Reads were aligned to the Genome Reference Consortium human genome build 38 (GRCh38), and the variants were annotated using Ensembl Variant Effect Predictor in combination with the in-house pipeline “ANATOLE2” (McLaren et al., 2016).
The identified variants were filtered based on their frequency, quality, and functional impact. Variants with a minor allele frequency greater than 1% in public databases (gnomAD) were excluded (Karczewski et al., 2020). We also removed variants with a Phred-scaled quality score of less than 30 and variants with a low read depth (<20). Variants located in genes known to be associated with the patient's phenotype according to the HPO database were given higher priority. Selected variants were confirmed by Sanger sequencing in the affected probands and their parents. The exons of the UGDH gene and the genetic variants were named according to the MANE transcript NM_003359.4, on the GRCh38 genome assembly. The nomenclature was validated using VariantValidator (Freeman et al., 2018). Primers are available on request.
2.2 Protein modeling
The structural model for the UGDH protein, chain A, was obtained from the RCSB Protein Data Bank (PDB) under the identifier 2Q3E (Berman, 2000; Rajakannan et al., 2011). To simulate the effect of specific mutations, the protein structure was modified using ChimeraX with the “swapaa” command (Meng et al., 2023). An analysis of the conservation of selected residues was performed by multiple sequence alignment using ClustalW, including the UniProt entries O60701, Q5R7B3, H2QPC9, A0A1D5Q620, P12378, O70475, O70199, and A8WGP7 (Sievers & Higgins, 2014). Missense effects on the 3D structure environment have been predicted using DynaMut2 for protein modeling (Rodrigues et al., 2021).
3 RESULTS
3.1 Clinical descriptions
The first patient, P1, is a 2-year-old male born to unrelated healthy parents, with no family history of neurodevelopmental disorders. He presented with severe developmental delay. He developed developmental epileptic encephalopathy in the neonatal period, characterized by abnormal movements, axial hypotonia, and dystonia. Despite several attempts, his epilepsy proved to be intractable. At 18 months of age, he was unable to support his head, although he maintained eye contact. He presented with microcephaly (−2SD). He had a normal brain magnetic resonance imaging (MRI) (Figure 1a) and metabolic report. This patient presented the common core phenotype characterized by severe developmental delay, epilepsy, and motor disorder with axial hypotonia.
The second patient, P2, is an 8-year-old female with global psychomotor delay. She achieved sitting at 13 months, standing at 18 months, and walking at 3.5 years. Her first words appeared at 13 months, but language regression was observed, and she used bisyllabic speech at 7 years of age. She also exhibited stereotypic behaviors, intolerance of frustration, self-aggression, and deficits in attention and concentration. Her growth parameters were normal (height—1DS, weight—2DS, head—0SD). She presented with an ataxic gait, scoliosis, convergent strabismus, and nystagmus. Patient P2 had an atypical phenotype, seizure-free with mild developmental delay and no facial dysmorphism. According to the patient's parents, there was no history of seizures, and a comprehensive evaluation by a pediatric neurologist revealed no signs suggestive of epilepsy. An electroencephalogram (EEG) could not be performed due to the patient's considerable agitation during the examinations. A cerebral MRI was performed and was considered unremarkable, showing no significant abnormalities (Figure 1b).
3.2 Identification of UGDH compound heterozygous variants
Trio exome sequencing analysis identified compound heterozygous variants in the UGDH gene in both patients. Patient 1 carried a compound association of NM_003359.4:c.[193C>T];[403C>T] p.[(Arg65*)];[(Arg135Trp)], and the patient 2 had the compound heterozygous variants NM_003359.4:c.[193C>T];[304C>T] p.[(Arg65*)];[(Arg102Trp)] (Figure 2a,b). The heterozygous nonsense variant p.(Arg65*) was reported at an extremely low allele frequency in gnomAD V4 (AF = 6.409e-5, no homozygous) (Chen et al., 2023), and resulted in a premature truncation in the NAD-binding domain. The nonsense variant was already known to be pathogenic (Hengel et al., 2020). Neither of the compound heterozygous associations was observed to co-occur in the gnomAD v2.1 database (Chen et al., 2023).
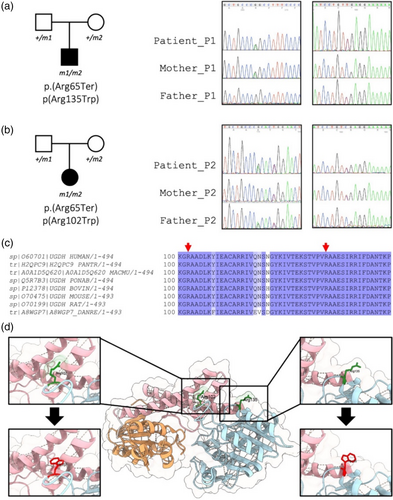
Both p.(Arg102Trp) and p.(Arg135Trp) missense variants affected highly conserved residues across species in the NAD-binding N-terminal domain of the UGDH protein (Figure 2c,d). These variants were rarely described in gnomAD V4, AF = 6.6e-6 and 6.7e-5, respectively, no homozygous reported, and involved a substitution of the highly hydrophilic residue, arginine, with the highly hydrophobic residue, tryptophan.
The missense variant p.(Arg102Trp) was associated with a CADD score of 33.0, a REVEL score of 0.715, and an AlphaMissense score of 0.496 (Ambiguous) (Cheng et al., 2023; Ioannidis et al., 2016; Kircher et al., 2014). A missense variant of uncertain significance involving the same arginine 102 with a different amino acid consequence p.(Arg102Gln) was reported in ClinVar (RCV003231884.1). The p.(Arg102Trp) variant was located in the N-terminal domain between the β4 and α5 secondary structures, within a disordered region of the NAD-binding domain (Figure 2d) (Rajakannan et al., 2011). The substitution could lead to a destabilizing effect of the region, with a ΔΔG score of −0.37 kcal/mol. The introduction of Trp102 would create new hydrophobic interactions between Tyr286 from the central domain and Trp102 (Figure 2d). The Arg102 residue is also known to be closed to Arg97 and Lys102 involved in the binding of UDP-Glc and/or UDP-GlcUA (Wang et al., 2019).
The p.(Arg135Trp) missense variant was predicted to be deleterious by in silico metapredictors, with high deleterious scores of 26.7, 0.732, and 0.741 for CADD, REVEL, and AlphaMissense (likely pathogenic), respectively (Cheng et al., 2023; Ioannidis et al., 2016; Kircher et al., 2014). Arg135 is the first amino acid of the α5 secondary structure (Rajakannan et al., 2011). The substitution could lead to a destabilizing effect of the area, with a ΔΔG score of −0.616 kcal/mol. The introduction of Trp135 would create new polar interactions between Thr244, from the central domain, and Trp135 (Figure 2d). A pathogenic missense variant, involving the same arginine 135, with a different amino acid consequence p.(Arg135Gln) has been described in a patient with severe refractory epilepsy infantile spasms, in a compound association with the missense variant p.(Ala24Val) (Shu et al., 2023).
4 DISCUSSION
In this report, we present the description of two patients with compound heterozygous variants in the UGDH gene. One patient exhibited the core phenotype of IDEE, whereas the other patient manifested global psychomotor delay and intellectual disability, without dysmorphic features or epilepsy (Table 1). The UGDH oxidoreductase enzyme consists of a complex association between trimer of UGDH dimers in a disk-like bilayer organization (Egger et al., 2011). Pathogenic human UGDH variants are thought to result in loss-of-function alleles, as indicated by previously published functional studies (Hengel et al., 2020; Shu et al., 2023).
| Phenotype | Patient 1 | Patient 2 | Literature |
|---|---|---|---|
| Developmental delay | + | + | 33/33 |
| Absence/delay of motor development | Absence | Delay | Absence 24/28, delay 4/28 |
| Absence/delay of speech | Absence | Delay | |
| Intellectual disability | Severe | Moderate | Severe 28/31, moderate 3/31 |
| Hypotonia | + | − | 32/32 |
| Epilepsy | + | − | 31/33 |
| Phamacoresistance | + | n/a | 18/21 |
| Feeding difficulties | + | − | 24/27 |
| Dysmorphic features | + | − | 20/26 |
| Brain MRI abnormalities | − | − | 13/23 |
No genotype–phenotype correlation was observed, and all the identified pathogenic missense variants are distributed throughout the UGDH gene without a hot spot and without any specific phenotypic features (Figure S1). To date, no patient is known to have biallelic nonsense variants, and no homozygous nonsense variants have been identified in the gnomAD database (Karczewski et al., 2020). The presence of a compound association of nonsense and missense variants does not appear to exacerbate the phenotype (Hengel et al., 2020; Shu et al., 2023). Since the first publication, three patients have been identified reinforcing the previously described core phenotype, which is briefly summarized in Table 1 (Hengel et al., 2020). Thus, the Jamuar syndrome is characterized by a severe developmental delay and hypotonia, including phenotypes from neonatal onset developmental epileptic encephalopathy (NDEE) to IDEE seizure spectrum. Notably, 3 of the 30 patients presented a mild phenotype with a motor disorder and seizures associated with fever. The onset of epilepsy occurred before 3 years of age (Hengel et al., 2020). IDEE has also been associated with compound heterozygous missense variants in two patients, namely p.[(Ala24Val)];[(Arg135Glu)] and p.[(Leu57Ile)];[(Thr244Lys)], who also presented with congenital microcephaly (Shu et al., 2023). This last description was consistent with patient P1, reported with the association p.[(Arg65*)];[(Arg135Trp)], confirming that mild microcephaly could be part of the phenotype.
The homozygous recurrent pathogenic missense variant p.(Arg317Gln) has been described in two patients with global developmental delay, and axial hypotonia, but without seizures at 5 years of age (Alhamoudi et al., 2020; Hengel et al., 2020). However, four other patients with the same homozygous missense variant have developed IDEE with onset between 12 and 30 months of age. Here, patient P2 with the association of the variants p.[(Arg65*)];[(Arg102Trp)] showed a severe phenotype without any episode of epilepsy. Interestingly, this patient presented with other rarely described clinical signs, such as ataxic gait, which was reported in only 2 of 30 patients, and abnormal eye movements (strabismus and nystagmus), which were not yet reported. The lack of epilepsy presentation in patient P2 could be explained by a residual expression of UGDH activity, as suggested by Hengel and collaborators to account for variability in disease severity (Hengel et al., 2020). The p.(Arg102Trp) variant has been predicted to modify intraprotein interactions with Tyr286 within the central domain. Tyr286 is adjacent to residues known to be associated with missense variants: Ile255, Gly271, and Met306 (Hengel et al., 2020). Tyr286 is located in the α11 helix, which contains the Asp280 and Cys276 residues. Both of these residues are involved in modifying the hexamer conformation and the interaction of the dimeric subunits (Rajakannan et al., 2011). We hypothesize that the p.(Arg102Trp) variant could indirectly affect homo-dimerization through changes in Tyr286 interactions, similar to what is suspected for other pathogenic missense variants, such as p.(Ile255Thr), p.(Gly271Arg), and p.(Met306Val) (Hengel et al., 2020).
The absence of homozygous nonsense variants in patients suggests that complete loss-of-function of the UGDH gene is likely lethal. Thus, complete gene knockout models in Drosophila, zebrafish, and mice have demonstrated the critical role of UGDH in cellular FGF signaling, developmental processes, and heart valve formation (Garcı́a-Garcı́a & Anderson, 2003; Superina et al., 2014; Walsh & Stainier, 2001). Notably, in vertebrate model organisms, complete knockout has consistently resulted in embryonic lethality, typically occurring around the gastrulation stage (Choksi et al., 2014; Garcı́a-Garcı́a & Anderson, 2003). In contrast to nonsense and splicing pathogenic variants, which likely lead to nonsense-mediated decay of the endogenous UGDH transcript, missense pathogenic variants are more likely to affect the stability of the enzyme and/or its oxidoreductase activity, as we may suspect for the two missense variants described here. Hengel and collaborators observed a significant decrease in the UGDH-catalyzed reduction of NAD+ to NADH in primary fibroblasts from patients with either the p.[(Arg393Trp)];[(Ala410Ser)], p.[(Tyr14Cys)];[(Ser72Pro)] associations or p.[(Ala82Thr)];[(Ala82Thr)] homozygous missense variants. In addition, cells from unaffected parents heterozygous for the p.(Ala82Thr) variant showed intermediate levels of NAD+ reduction, and cells from patients with homozygous p.(Ala82Thr) variants showed a reduction in hyaluronic acid synthesis, a process requiring UDP-glucuronate, a product of UGDH enzymatic activity. This suggests that the severity of epileptic encephalopathy may correlate with the amount of residual UGDH activity. Sufficient UGDH activity may be necessary for gastrulation to occur during early human embryonic stages, but may be limiting for neuronal development thereafter (Hengel et al., 2020).
Several diseases involving proteins of glycosaminoglycan metabolism proteins have been described. Biallelic alteration of the NDST1 gene, encoding a bifunctional GlcNAc N-deacetylase/N-sulfotransferase, has recently been proposed as a candidate gene for autosomal recessive intellectual disability in two families, with muscular hypotonia, epilepsy, and postnatal growth failure (Reuter et al., 2014). In mice, Ndst1 is considered essential for embryonic development, and homozygous null variants are lethal during the perinatal period (Reuter et al., 2014). The UDP-glucose pyrophosphorylase (UGP2) gene, which encodes an octameric enzyme involved in the nucleotide sugar metabolism, has also been implicated in patients with IDEE. As observed in patient P1 from the paper, biallelic UGP2 pathogenic variants were associated with a severe form of intractable epilepsy, severe developmental delay, microcephaly, visual impairment, and similar minor dysmorphisms (Perenthaler et al., 2020).
Comparisons with other disorders involving glycosaminoglycan metabolizing proteins highlight the complexity of these conditions. Our findings contribute to the phenotype of UGDH-related disorders description and highlight the need for further research to elucidate the intricate genotype–phenotype relationships in these rare genetic conditions.
AUTHOR CONTRIBUTIONS
P.B.P. and T.S. wrote the manuscript. P.B.P., S.B., M.R., P.B., C.T., and T.S. performed the experiments and the data interpretation. E.A.Y. performed the bioinformatics analysis. C.V., R.C., C.C., and J.G. collected and evaluated the clinical and genetic data. T.S. and J.G. revised the manuscript. All authors discussed the results, commented on the manuscript, and approved the final manuscript.
ACKNOWLEDGMENTS
We thank the patients and their families for participating in this study.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




