Unraveling the molecular diagnosis of metaphyseal enchondromatosis with D-2-hydroxyglutaric aciduria: A 22-year quest
To the Editor,
Metaphyseal enchondromatosis with D-2-hydroxyglutaric aciduria (MC-HGA) (OMIM 614875) is characterized by chondrodysplasia and the presence of urinary excretion of D-2-hydroxyglutaric acid. Vascular anomalies have been described in previous studies (Choo et al., 2012; Spranger et al., 1978; Yeetong et al., 2018). It is associated with somatic point variants in IDH1 (OMIM*147700) including c.394C>A p.(R132S) and c.395G>A p.(R132H) (Vissers et al., 2011). However, the underlying variants in many patients have not been identified (Corvino et al., 2022). Here, we present a 22-year journey to provide a molecular diagnosis for a patient with MC-HGA and offer recommendations for molecular diagnosis based on our experience.
A 22-year-old Thai patient with MC-HGA presented with metaphyseal enchondromatoses first observed at 11 months of age and hemangiomas that started at 3 years of age, along with D-2-hydroxyglutaric aciduria, as previously reported (Yeetong et al., 2018). The enchondromas involved all long bones and were symmetrical and progressively enlarged. The hemangiomas increased in number and size, affecting the skin and mucosa throughout the body, including the scalp, eyelids, ala nasi, larynx, trachea, lips, tongue, buccal mucosa, hands, and feet (Figure 1a). These hemangiomas led to repeated episodes of bleeding during haircuts and while eating, as well as causing breathing difficulties, wheezing, and challenges in walking.
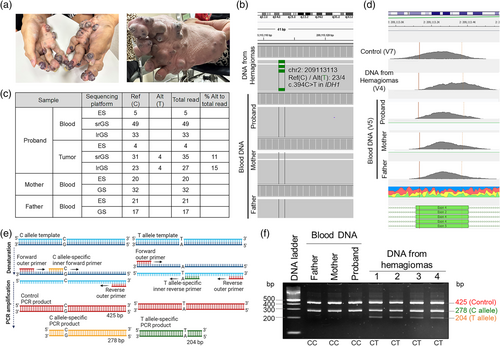
To identify the causative variant, targeted Sanger sequencing of genes involving in enchondromatosis and D-2-hydroxyglutarate metabolism including IDH1, PTHR1, D2HGDH, and HOT using DNA extracted from leukocytes, and trio exome sequencing (ES) were performed in 2014 using DNA extracted from hemangiomas and leukocytes, but were unsuccessful (Yeetong et al., 2018) Considering the possibility of non-coding or structural somatic variants, we performed short-read genome sequencing (srGS) using Illumina's Novaseq 6000 and HiFi long-read GS (lrGS) using PacBio Sequel IIe. Regular bioinformatic pipeline did not yield any pathogenic variants. However, reanalysis by scrutinizing the BAM files with a focus on IDH1 and IDH2 identified few reads with the c.394C>T p.(R132C) in IDH1 (NM_005896.4, Clinvar SCV004028549) in both srGS (Alt:Ref = 4:31; 11%) and lrGS (Alt:Ref = 4:23; 15%) in the proband's hemangioma. Not a single read with the c.394C>T variant was found in blood samples of the patient or his parents (Figure 1b,c). To the best of our knowledge, this represented the first identification of the c.394C>T p.(R132C) variant in the IDH1 gene in a patient with MC-HGA.
Retrospectively, this variant could not be identified previously by srGS and lrGS with the regular analytic pipeline because the read depth threshold for the presence of a heterozygous alternate allele was generally set at ≥20%, while the somatic mosaic variant was found in ≤15%. Moreover, the variant was not found by ES because the enrichment reagent and the sequencing method we used for ES at that time, provided insufficient read depth at the end of exon 4 of IDH1, where the variant lies (Figure 1c,d).
To confirm the somatic mosaic variant in the proband's hemangiomas, we conducted tetra-primer amplification refractory mutation system-polymerase chain reaction (T-ARMS-PCR) (Medrano & de Oliveira, 2014). All tested samples provided a 425-bp control band and a 278-bp band representing the wild-type allele. In addition to these two bands, the four independent hemangioma samples showed a 204-bp band, indicating the presence of the somatic mutant c.394C>T allele in IDH1 (Figure 1e,f). As the variant was present in multiple hemangiomas but absent in leukocytes, we hypothesized that it occurred in specific mesodermal cells during development.
Recent clinical trials have shown promising results with ivosidenib, an inhibitor of the mutant IDH1 enzyme, and vorasidenib, a dual inhibitor targeting both IDH1 and IDH2. These inhibitors have demonstrated a reduction in tumor levels of D-2-hydroxyglutarate, increased DNA 5-hydroxymethylcytosine, decreased tumor cell proliferation, and suppressed immune cell activation in patients with mutant IDH1 glioma (Mellinghoff et al., 2023). These inhibitors hold potential as new therapies for patients with MC-HGA and hemangioma who exhibit the IDH1 variants.
Drawing from the lessons learned in this patient, we recommend using DNA extracted from tumors for molecular diagnosis in patients with MC-HGA and conducting deep sequencing with a low read depth threshold. Manual analysis of the BAM file may be necessary. Finally, T-ARMS-PCR is a sensitive technique for confirming the presence of low-percentage somatic mosaic variants. Importantly, accurate detection of pathogenic IDH1 variants is crucial for guiding personalized drug therapy for patients.
AUTHOR CONTRIBUTIONS
Khanti Rattanapornsompong, Thantrira Porntaveetus, Vorasuk Shotelersuk: Study design, formal analysis, writing—original draft. Wanna Chetruengchai, Chalurmpon Srichomthong, Thanakorn Theerapanon: Formal analysis. All authors: Writing—review and editing.
FUNDING INFORMATION
This project was funded by the Health Systems Research Institute (66-101, 66-122), National Research Council of Thailand (N42A650229), Faculty of Dentistry (DRF67_012), Faculty of Medicine (RA-MF-08/66), and Thailand Science Research and Innovation Fund Chulalongkorn University. KR was supported by Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
PATIENT CONSENT STATEMENT
Written informed consent was obtained from the patient to publish clinical data and images.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.