Probable digenic inheritance of Diamond–Blackfan anemia
Abstract
A 26-year-old female proband with a clinical diagnosis and consistent phenotype of Diamond–Blackfan anemia (DBA, OMIM 105650) without an identified genotype was referred to the Undiagnosed Diseases Network. DBA is classically associated with monoallelic variants that have an autosomal-dominant or -recessive mode of inheritance. Intriguingly, her case was solved by a detection of a digenic interaction between non-allelic RPS19 and RPL27 variants. This was confirmed with a machine learning structural model, co-segregation analysis, and RNA sequencing. This is the first report of DBA caused by a digenic effect of two non-allelic variants demonstrated by machine learning structural model. This case suggests that atypical phenotypic presentations of DBA may be caused by digenic inheritance in some individuals. We also conclude that a machine learning structural model can be useful in detecting digenic models of possible interactions between products encoded by alleles of different genes inherited from non-affected carrier parents that can result in DBA with an unrealized 25% recurrence risk.
1 INTRODUCTION
Diamond–Blackfan Anemia (DBA, OMIM 105650) is characterized by the failure of red blood cell production as well as congenital anomalies, and cancer predisposition with variable expressivity and reduced penetrance (Jahan et al., 2020). In most cases, DBA is characterized as an autosomal dominant ribosomopathy caused by loss-of-function heterozygous variants in one of 19 ribosomal protein (RP) genes, that encode the following proteins of either the small (RPS19, RPS26, RPS10, RPS24, RPS17, RPS7, RPS27, RPS29, RPS28, RPS15A) or the large (RPL5, RPL11, RPL35A, RPL26, RPL15, RPL31, RPL27, RPL35, RPL18) ribosomal subunits (Ulirsch et al., 2018).
Clinical presentations of DBA have variable expressivity and reduced penetrance that result in a broad spectrum of phenotypes that range from mild to severe. In addition to anemia, ~30%–50% of patients with DBA present with pleiotropic congenital abnormalities including craniofacial, upper limb, heart, genitourinary, and ophthalmologic congenital malformations, as well as microcephaly, growth failure, and short stature (Da Costa et al., 2010; Landowski et al., 2013; Vlachos & Muir, 2010). Vlachos and Muir (2010) reported that the abnormalities commonly observed included craniofacial (50%), skeletal (39%), mostly upper limb and hand, genitourinary (38%), and cardiac (30%). Ophthalmologic abnormalities such as congenital glaucoma and cataract can also be seen. RPS19 variants that cause DBA can be associated with variable phenotypic expressivity, and the genotype–phenotype correlation in patients with RPS19 variants is poorly understood. Gazda et al. (2008) have reported that RPL5 and RPL11 variants were more frequently associated with congenital abnormalities than were RPS19 variants. Wang et al. (2015) identified a de novo variant in RPL27 in a patient with DBA who presented with an atrial septal defect and pulmonary stenosis in the absence of a family history of anemia. To our knowledge, that RPL27 variant is the only previous report of a putatively causal RPL27 variant in DBA, and Wang et al. implied a la Occam's razor that the single RPL27 variant was sufficient to explain the DBA in their patients.
Among the RP causing genes, it is reported that ~25% of DBA cases are associated with RPS19 gene variants, which are reported to be the most common single gene cause of DBA (Gazda et al., 2008). In general, DBA results from single, monoallelic variants in RP genes that have an autosomal dominant mode of inheritance. Most DBA cases are sporadic, but ~10%–25% are familial and most have an autosomal dominant mode of inheritance (Willig et al., 1999). An autosomal-recessive inheritance pattern has also been reported (Gustavsson et al., 1997). A few cases of DBA have been reported to have a possible digenic inheritance with two non-allelic variants such as RPL5 and RPS24 (Quarello et al., 2010), but this has not been proven.
We report here an unusual case of DBA caused by digenic inheritance that could not be explained by a simple Mendelian mode of inheritance. In this case, we predicted the digenic interaction between the RPS19 and RPL27 variants by a structural biological model using machine learning, which was subsequently supported by co-segregation analysis and RNA sequencing.
2 CASE REPORT
Our patient is a 26-year-old female who has a history of multiple problems in infancy and childhood including anemia, glaucoma, microcephaly, atrial septal defect, multicystic and dysplastic kidneys, scoliosis, short stature, growth hormone deficiency, and Vitamin D deficiency. She has presented with chronic constitutional red cell aplasia with macrocytic erythrocytes, anemia, and reticulocytopenia. Her family history revealed that her parents and six sibs (including three brothers and three sisters) were all in good health and none had any findings of DBA. Her repeated physical exams were notable for short stature and microcephaly (head circumference <3 percentile). No cataracts were noted. Her inner intercanthal, outer intercanthal, and interpupillary distances were all <3 percentile, but her palpebral fissure length was within normal limits. Scoliosis and genu valgus were also noted. Her neurologic examinations were normal. Neither of her parents or sibs had any phenotypic features of DBA.
She had chromosomal microarrays done at 8 and 16 years of age, and both were reported as normal. She had DNA sequencing and deletion/duplication analysis of her RPS19 genes at 14 years of age. Her DNA results showed heterozygosity for a p.Thr55Met RPS19 likely benign variant that was shared with her nonaffected mother. At 16 years of age, because she and her unaffected mother shared the same p.Thr55Met RPS19 likely benign variant, RPL11 sequencing was done to detect genocopies of DBA and the results were negative. Her studies for chromosomal breakage were negative. Her sequencing of CREBBP gene for Rubinstein–Taybi syndrome as well as her 7-dehydrocholesterol test for Smith–Lemli–Opitz syndrome were both negative.
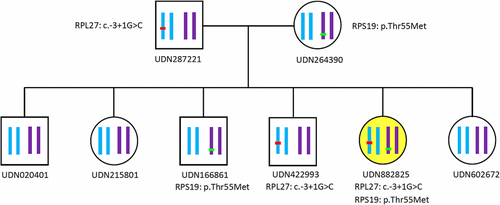
In summary, while she was clinically diagnosed with DBA due to the strength of her phenotype, the diagnosis was not genetically confirmed. As such, she was referred to the Undiagnosed Diseases Network (UDN) and enrolled at the Vanderbilt UDN clinical site to identify a genetic cause. Her UDN exome sequencing (ES) obtained at 21 years of age confirmed that she was heterozygous for a p.Thr55Met RPS19 variant of unknown significance that was inherited from her unaffected mother. Importantly, her subsequent UDN genomic sequencing (GS) obtained at the age of 21 revealed that she was also heterozygous for a non-coding c.-3+1G>C RPL27 (NM_000988.5) variant that had been missed by ES. The exon impacted by the variant codes for 5′UTR sequence (see Supplementary Figure 1) (den Dunnen et al., 2016). This variant is predicted to perturb mRNA splicing and to be likely pathogenic and was inherited from her unaffected father (see Figure 1). No abnormal variant in the other RP genes was found.
Her diagnosis of DBA was further supported by her hemoglobin F level, which was elevated at 2.9% (reference range: 0%–2%). Also, her erythrocyte adenosine deaminase level, which was slightly below normal at 390 mU/kg Hb (reference range: 400–900 mU/g Hb).
METHODS AND RESULTS
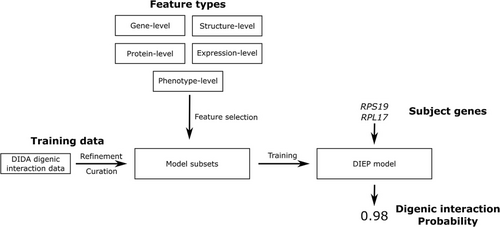
To investigate the possibility of a digenic interaction between her RPS19 and RPL27 variants, we conducted a computational structural biology analysis using the Digenic Interaction Effect Predictor (DIEP) software (Yuan et al., 2022) (see Figure 2). The DIEP software is a machine-learning-based method capable of identifying digenic and pseudo-digenic effects between gene pairs. Building on an existing method developed as part of our UDN efforts (Mukherjee et al., 2021), DIEP was trained on 140 known digenic pairs from the Digenic Diseases DAtabase (DIDA) (Gazzo et al., 2016) which served as the positive set, along with ~16,000 gene pairs generated randomly or identified to lack digenic effects by the DIDA database, which served as the negative set. Gene-, protein-, structure-, expression-, and phenotype-level features extracted from the genetic dataset were used to train the algorithm, which was then used to calculate a large number of digenic interactions wherein a result of 1 indicates very strong digenic interactions, and 0.5 is considered as the significance cutoff. This model therefore allows gene level data (e.g., two genotypes) digenic probabilities to be estimated based on multilevel training data. Our gene (one level) calculations comparing RPS19 and RPL27 yielded a score of 0.98, which strongly suggests a digenic interaction between the RPS19 and RPL27 variant products. These results combined with variants located at different loci, suggested that her atypical phenotypic presentation of DBA was caused by a digenic inheritance pattern.
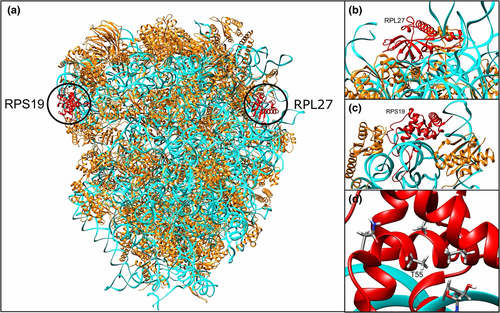
To further investigate our hypothesis that a digenic gene interaction between genes caused DBA, we inspected the different 80S ribosome structures available in the Protein Data Bank (PDB) (Burley et al., 2022), and in all of them, RPS19 and RPL27 are on opposite sides of the complex and do not make direct contact (see Figure 3). This model also suggests that altered amino acid Met in the 55 position of RPS19 may make contact with the ribosome and this change in amino acid (from Thr) could be responsible for the loss of function. This suggests that the predicted digenic effect should be caused by an indirect interaction between the two proteins mediated through ribosome function rather than a direct contact between them.
To further strengthen our hypothesis that this is digenic inheritance, we conducted co-segregation analysis of her maternal p.Thr55Met RPS19 variant and her presumed paternal c.-3+1G>C RPL27 variant. To do this, genomic DNA was extracted from whole blood of the proband, both her parents and all her sibs. These DNA samples were used as PCR templates to amplify segments of the genes RPS19 (NM_001022.4) and RPL27 (NM_000988.5) that contained the variants. The primer pairs used were: RPS19_forward (5′-CTGTAAAAGACGTGAACCAGCAG-3′), RPS19_reverse (5′-AGGACACACAGCCAGATGTAAAA-3′), RPL27_forward (5′-CTTCTTTCCTTTTTGCTGGTAGGG-3′), and RPL27_reverse (5′-TCTCAAAAACTCTCCCACGTTCA-3′). PCR products were subjected to Sanger DNA sequencing. The results showed that the proband's c.-3 + 1G > C RPL27 variant was inherited from her heterozygous father and that while one brother was heterozygous for the RPL27 variant and another brother was heterozygous for the proband's p.Thr55Met maternal RPS19 variant that none shared both of these variants (see Figure 1). These results confirmed that the proband, but none of her sibs, was a compound heterozygote for her prenatal RPS19 and RPL27 variants, supporting the theory of digenic interaction as being the cause of her DBA.
To further our understanding of the impact of these variants on expression, we conducted RNA sequencing on the proband's whole blood total RNA (using PAX gene tubes and the manufacturer's instructions) and analyzed both variants. For the heterozygous RPS19 variant (NM_001022.4 p.Thr55Met), no changes in splicing were detected and the expression level of the transcript was within normal range compared to controls. Analysis of the RPL27 transcripts heterozygous for the variant (NM_000988.5 c.-3+1G>C) demonstrated 50% reduced expression compared to a reference dataset of 202 control samples indicating haploinsufficiency. A very low-level transcript containing retention of Intron 1 was also detected, not present in the control samples, and is most likely due to the c.-3+1G>C Intron 1 variant. Addition of these extra 277 bases in Intron 1 into the 55 bp 5′UTR appears to make the transcript unstable and result in RPL27 transcript haploinsufficiency.
3 DISCUSSION
Here we describe a patient with a phenotype of DBA caused by a likely digenic interaction between RPS19 and RPL27 variants that was predicted by a machine learning algorithm and confirmed with co-segregation analysis. To our knowledge, there is no previous report of cases of DBA caused by a digenic effect of two non-allelic variants that was evaluated by similar structural biology analysis.
In general, DBA is associated with single, monoallelic variants in RP causing genes. None the less, we felt at her initial UDN evaluation that her atypical DBA phenotype was not fully explained by her RPS19 variant that she shared with her non-affected mother. In addition, most DBA cases are reported to be sporadic due to de novo mutations and only ~10%–25% are familial, most of which show autosomal dominant inheritance (Willig et al., 1999). Variable expressivity was considered as a possible explanation of why her mother shared her RPS19 variant, but no clinical features of DBA were detected on clinical assessment of her mother (Carlston et al., 2017).
Intriguingly, a non-coding presumed paternal RPL27 variant that had been missed by ES was detected by GS demonstrating its clinical utility. Based on detection of her RPL27 variant, we developed a hypothesis that her atypical clinical phenotypic presentation of DBA may be due to possible digenic inheritance between her RPS19 and RPL27 variants (see Figure 1). While digenic inheritance has been postulated to result in DBA (Quarello et al., 2010), here we demonstrate this phenomenon in a patient. Further study is required to identify if a possible digenic effect might explain previously published variable expressivity (Carlston et al., 2017).
Digenic inheritance is a simple genetic model in which affected individuals are compound or double heterozygotes for variants in two different genes that encode different proteins. Kajiwara et al. reported a case of retinitis pigmentosa as the first case of a digenic inheritance in human disease (Kajiwara et al., 1994). Since then, multiple digenic diseases have being discovered. Forty-four different digenic diseases were registered in the DIDA in 2016 (Gazzo et al., 2016). Digenic inheritance has been proposed to describe a clinical presentation that is not able to be completely explained by a simple autosomal-dominant or -recessive mode of inheritance. For example, although more than half of all UDN cases remain undiagnosed, Mukherjee et al. hypothesizes that many of these unsolved, rare genetic diseases might be caused by multiple variants in more than one gene as a result of digenic inheritance (Mukherjee et al., 2021). The DIEP score of 0.98 indicated the presence of a very likely digenic interaction between the genes RPS19 and RPL27, consistent with data shown in Figures 1 and 2.
In summary, we describe a case of atypical phenotypic DBA predicted to be caused by digenic interactions between RPS19 and RPL27 variant products predicted by structural biology analysis, co-segregation, and RNA sequencing. We suggest that atypical phenotypic presentations of DBA may be caused by digenic inheritance pattern in some individuals with DBA. We also conclude that structural biology modeling can be useful in detecting digenic models of possible interactions between products encoded by alleles of different genes inherited from non-affected carrier parents that can result in DBA with atypical phenotypic presentations, but which have an unrealized 25% recurrence risk.
AUTHOR CONTRIBUTIONS
Yutaka Furuta drafted the article. John A. Phillips III, Rizwan Hamid, Joy D. Cogan, Lynette Rives, Alican Gulsevin, Qi Liu, and Hua-Chang Chen were responsible for the correct description display and interpretation of the genomic diagnostics and technical parts. Alican Gulsevin, Lynette Rives, and Joy Cogan helped to create figures. Rory J. Tinker, Serena M. Neumann, John A. Phillips III, Rizwan Hamid, Joy Cogan, Lynette Rives, Alican Gulsevin, and Karen M. Joos helped to draft the article. All authors read, corrected and approved the final article.
ACKNOWLEDGMENTS
The authors are grateful to the patient for participating in the UDN. This work was supported in part by the National Institutes of Health (NIH) Common Fund, NIH/NHGRI grant 3U01NS134349 (J.A.P., J.H.N., and R.H.). Consortia: Collaborators of the Undiagnosed Disease Network (UDN) include Maria T. Acosta, David R. Adams, Raquel L. Alvarez, Justin Alvey, Aimee Allworth, Ashley Andrews, Euan A. Ashley, Ben Afzali, Carlos A. Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Hugo J. Bellen, Jimmy Bennett, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Lauren C. Briere, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Olveen Carrasquillo, Thomas Cassini, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Ivan Chinn, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, Matthew Coggins, F. Sessions Cole, Heather A. Colley, Heidi Cope, Rosario Corona, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D'Souza, Hongzheng Dai, Surendra Dasari, Joie Davis, Jyoti G. Dayal, Margaret Delgado, Esteban C. Dell'Angelica, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L. Doss, Emilie D. Douine, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Marni Falk, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, Jiayu Fu, William A. Gahl, Ian Glass, Page C. Goddard, Rena A. Godfrey, Alana Grajewski, Andrea Gropman, Meghan C. Halley, Rizwan Hamid, Neal Hanchard, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yan Huang, Sarah Hutchison, Wendy Introne, Rosario Isasi, Kosuke Izumi, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Emerald Kaitryn, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N. Kobren, Isaac S. Kohane, Jennefer N. Kohler, Susan Korrick, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Seema R. Lalani, Byron Lam, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, Kimberly LeBlanc, Brendan H. Lee, Roy Levitt, Richard A. Lewis, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, AudreyStephannie Maghiro, Rachel Mahoney, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Alexa T. McCray, Elisabeth McGee, Matthew Might, Danny Miller, Ghayda Mirzaa, Eva Morava, Paolo Moretti, Marie Morimoto, John J. Mulvihill, Mariko Nakano-Okuno, Stanley F. Nelson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Jeanette C. Papp, Neil H. Parker, Leoyklang Petcharet, John A. Phillips III, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey Swerdzewski, Aaron Quinlan, Deepak A. Rao, Anna Raper, Wendy Raskind, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Elizabeth Rosenthal, Francis Rossignol, Maura Ruzhnikov, Marla Sabaii, Ralph Sacco, Jacinda B. Sampson, Mario Saporta, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, Elaine Seto, Prashant Sharma, Vandana Shashi, Emily Shelkowitz, Sam Sheppeard, Jimann Shin, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Edward C. Smith, Kevin S. Smith, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C. Spillmann, Andrew Stergachis, Joan M. Stoler, Kathleen Sullivan, Jennifer A. Sullivan, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Queenie K.-G. Tan, Amelia L. M. Tan, Arjun Tarakad, Herman Taylor, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Rachel A. Ungar, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Melissa Walker, Nicole M. Walley, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Monika Weisz Hubshman, Mark Wener, Tara Wenger, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Kim Worley, Shinya Yamamoto, Zhe Zhang, and Stephan Zuchner.
CONFLICT OF INTEREST STATEMENT
Th authors declare no conflicts of interest.
DATA AVAILABILTY STATMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.




