Indian patients with CHST3-related chondrodysplasia with congenital joint dislocations
Abstract
CHST3-related chondrodysplasia with congenital joint dislocations (CDCJD, #MIM 143095), is a rare genetic skeletal disorder caused by biallelic loss of function variants in CHST3. CHST3 is critical for the sulfation of chondroitin sulfate. This study delineates the clinical presentation of nine individuals featuring the key symptoms of CDCJD; congenital joint (knee and elbow) dislocations, short trunk short stature progressive vertebral anomalies, and metacarpal shortening. Additional manifestations include irregular distal femoral epiphysis, supernumerary carpal ossification centers, bifid humerus, club foot, and cardiac abnormalities. Sanger sequencing was carried out to investigate molecular etiology in eight patients and exome sequencing in one. Genetic testing revealed five homozygous variants in CHST3 (four were novel and one was previously reported). All these variants are located on sulfotransferase domain of CHST3 protein and were classified as pathogenic/ likely pathogenic. We thus report on nine individuals with CHST3-related chondrodysplasia with congenital joint dislocations from India and suggest monitoring the health of cardiac valves in this condition.
1 INTRODUCTION
CHST3-related chondrodysplasia with congenital joint dislocations (CDCJD, #MIM 143095), is an autosomal recessive chondrodysplasia. It belongs to the sulphation disorder group (group 4) in nosology of skeletal genetic disorders: 2023 revision (Unger et al., 2023). CHST3-related CDCJD is characterized by congenital joint dislocations (predominantly elbows, hips, or knee joints), short stature of prenatal onset, progressive kyphosis/scoliosis, progressive vertebral defects, platyspondyly and club foot (Unger et al., 2010).
CHST3-related CDCJD is caused by defects in sulphation metabolism of proteoglycan. Biallelic disease-causing variants in CHST3 (carbohydrate sulfotransferase 3) result in the deficiency of sulfotransferase activity. CHST3 encodes for chondritin-6-sulfotranferase, an enzyme that catalyzes transfer of sulfate group from 3′-phospho-5′-adenylyl sulfate (PAPS) to position 6 of chondroitin sulfate. Chondroitin sulfate is a proteoglycan present in cartilage and distributed on the surface of most cells and extracellular matrices (Thiele et al., 2004). Hence, insufficient sulphation by CHST3 gene at a 6′ position of proteoglycan present cartilage result in a spectrum of symptoms which collectively described as CDCJD-CHST3 type. Exact prevalence of CHST3-related CDCJD is not known. However, till date, nearly 62 individuals affected with CHST3 deficiency are reported (Duz & Topak, 2020; Kausar et al., 2022).
CHST3 gene has three exons, of which only exon 2 and exon 3 are coding. Hence, if the diagnosis is definitive then testing CHST3 gene by conventional PCR followed by Sanger sequencing can be cost-effective, accurate, and can provide a rapid molecular diagnosis for patients in Indian settings. Here, we describe clinical, radiological, and molecular findings of nine individuals affected with CHST3-related CDCJD from five Indian families that would enable clinic-radiological and molecular diagnosis of similarly affected patients.
2 MATERIALS AND METHODS
2.1 Patients
We reviewed nine individuals (P1–P9) affected with CHST3 deficiency from five families of Indian origin in this study. Family history, clinical, and radiological findings of all with CHST3-related CDCJD were noted, and written informed consents from the affected individuals and their family members were obtained. 2-3 mL of EDTA blood sample was collected from probands, siblings, and their parents. Genomic DNA extraction was performed using the standard protocol. The study was approved by the Institutional Ethics Committee, Kasturba Medical College, Manipal.
2.2 Sanger sequencing for single gene testing
Single gene testing was performed for the probands, P3, P4, P5, and P7 from four families. The primers for coding exons 2 and 3 along with their flanking exon–intron boundaries of CHST3 (NM_004273.5) were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). Primers are available on request. Polymerase chain reaction (PCR) amplification was performed using genomic DNA obtained from the proband and their parents blood samples. Sanger sequencing was done using the purified PCR products in ABI 3500 genetic analyzer. The disease-causing variants identified in the probands were further segregated in the affected siblings, and their parents (Figure S2).
2.3 Exome sequencing
Exome sequencing (ES) was performed in one patient (P1) from family 1, as the clinical diagnosis was not apparent. ES was carried out using MGIEasy Exome Capture V5 Probe Set (Shenzhen, China). Annotation and analysis of exome data were conducted in accordance with our in-house protocol. In brief, the raw data was aligned to GRCh38 assembly using Burrows–Wheeler Aligner (v0.7.15) and our in-house pipeline using Genome Analysis Toolkit Best Practices. ANNOVAR (Wang et al., 2010) and customized in-house scripts were used for the annotation (Kausthubham et al., 2021). Sanger sequencing was done for validation and segregation analysis of the identified variant in his brother and parents.
Pathogenicity of all variants mentioned in this study was evaluated using in silico prediction tools, CADD phred, REVEL, and M-CAP. Frequency of reported variants were assessed in in-house data of 2632 exomes and control population database, gnomAD. All the identified variants in this study were represented using HGVS nomenclature and classified according to American College of Medical Genetics and Genomics (ACMG) 2015 criteria (Richards et al., 2015). All these variants were submitted to ClinVar database.
3 RESULTS
3.1 Clinical and radiographic findings
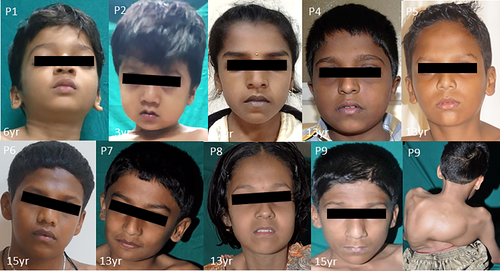
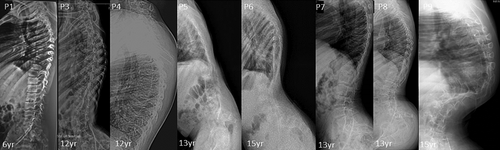
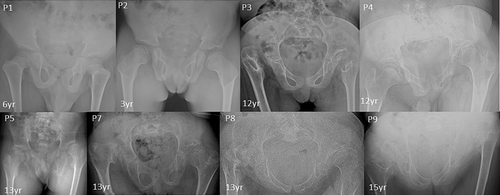
Seven male and two female patients were clinically diagnosed with CDCJD from five unrelated families of Indian origin. Four of these families were noted to be consanguineous (Figure S1). The age at examination ranged from 3 to 15 years. The main presenting clinical features associated with the condition were joint dislocations [9/9; elbow joint dislocation/contractures (7/9), hip dislocation (5/9)], short stature (9/9), progressive kyphosis (8/9), club foot (7/9) and hallux valgus (5/9). Genu valgum (4/9), pectus carinatum (3/9), camptodactyly (2/9), and pes planus (1/9) are the rare phenotypic features observed in our cohort.
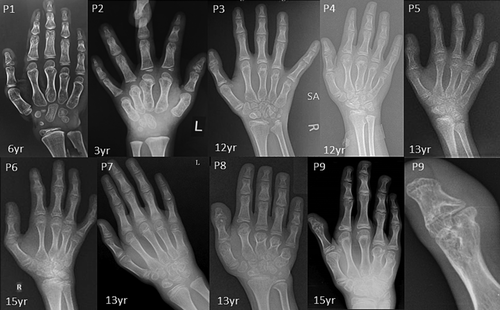
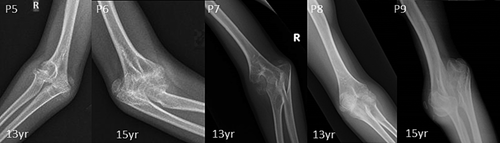
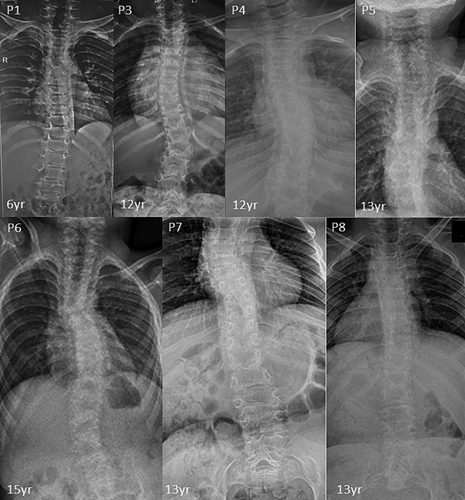
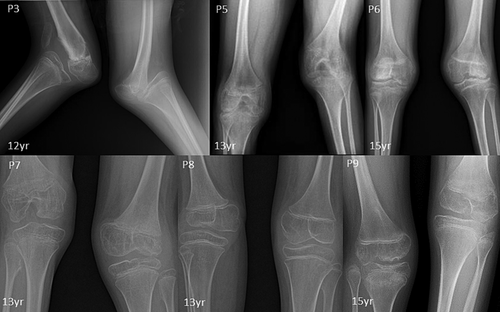
Radiographic features including shortened 4th and 5th metacarpals (9/9), flattened proximal femoral epiphysis (9/9), supernumerary carpal bones (8/9), scoliosis (7/9), coxa vara (6/9), widened inter condylar notch (6/9), distal bifid humeri (5/9), short femoral neck (4/9), and platyspondyly (4/9), were most commonly observed. Bifid distal end of proximal phalange and proximal end of distal phalange of thumb can be noted in three individuals. Increase in interpedicular distance from T12 to L2 and unossified ulna was clearly evident in P7. Along with shortened 4th and 5th metacarpals, 3rd metacarpal was also shortened in P8.
Cardiac anomalies such as atrioventricular valve prolapse (1/9), mitral valve thickening, bicuspid aortic valve, and mild tricuspid regurgitation (3/9) were noted. No significant intellectual disability, facial dysmorphism and hearing loss were observed in our cohort. Detailed clinical and radiological findings are outlined in Table 1, and Figures 1-7.
| Family ID | Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Subject ID | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 |
| Subject characteristics | |||||||||
| Age at examination | 6 years | 3 years | 12 years | 13 years | 13 years | 15 years | 13 years | 13 years | 15 years |
| Gender | Male | Male | Female | Male | Male | Male | Male | Female | Male |
| Consanguinity | − | − | + | + | + | + | + | + | + |
| Ethnicity | Asian Indian | Asian Indian | Asian Indian | Asian Indian | Asian Indian | Asian Indian | Asian Indian | Asian Indian | Asian Indian |
| Height in cm (SD) | 100(−3.64) | 80.5(−4.10) | NA | Could not stand | 97(−7.60) | NA | 104(−6.70) | 92(−9) | 101(−8.32) |
| Weight in kgs (SD) | 15(−2.87) | 10(−3.17) | NA | 29.8 (−2.32) | 20.86 (−3.64) | NA | 18(−4.06) | 15(−4.62) | 20(−4.56) |
| Head circumference in cm (SD) | 49.5(1.72) | 47.5 (−1.28) | NA | 50 (−2.82) | 51(−2.15) | NA | 49(−3.49) | 50 (−3.76) | 52(−1.95) |
| Arm span | 21 | 12 | NA | NA | 110 | NA | 109 | 93 | 102 |
| Lower segment (cm) | 53 | 49.5 | NA | NA | 53 | NA | 51 | 43 | 49 |
| Clinical features | |||||||||
| Short stature | + | + | + | + | + | + | + | + | + |
| Mesomelia | − | − | − | − | − | − | + | + | + |
| Rhizomelia | + | − | + | NA | − | + | − | − | − |
| Pectus Carinatum | NA | NA | NA | − | − | − | + | + | + |
| Camptodactyly | − | − | − | + | + | + | − | − | − |
| Hallux valgus | − | NA | − | − | + | + | + | + | + |
| Echocardiography | NA | NA | NA | NA | NA | NA | + | + | + |
| Genu valgum | + | + | − | NA | + | + | + | − | − |
| Club foot | − | − | + | + | + | + | + | + | + |
| Radiological features | |||||||||
| Platyspondyly | + | − | − | − | − | − | + | + | + |
| Kyphosis | + | − | + | + | + | + | + | + | + |
| Scoliosis | + | − | + | + | + | + | + | + | + |
| Coronal clefts | + | − | − | − | − | − | + | + | − |
| Increased interpedicular distance (T12-L1/2) | − | − | − | − | − | − | + | + | + |
| 4th and 5th shortened metacarpal bones | + | + | + | + | + | + | + | + | + |
| Irregular and accessory carpal ossification center | − | − | + | + | + | + | + | + | + |
| Bifid humerus | NA | NA | NA | NA | + | + | + | + | + |
| Elbow dislocation/dysplasia | + | + | NA | NA | + | + | + | + | + |
| Knee joint dislocation/ dysplasia | − | + | + | − | − | − | − | − | − |
| Flattened proximal femoral epiphysis | + | + | + | + | + | + | + | + | + |
| Short femoral neck | − | − | − | + | − | − | + | + | + |
| Widened inter condylar notch | − | NA | + | NA | + | + | + | + | + |
| Hip joint dysplasia/dislocation | − | − | + | + | − | − | + | + | + |
| Coxa vara | − | − | + | NA | + | + | + | + | + |
| Epiphyseal dysplasia | + | + | + | + | + | + | + | + | + |
| Extra skeletal findings | |||||||||
| Hearing impairment | − | − | − | − | − | − | − | − | − |
| Cardiac involvement | − | − | + | − | − | − | + | + | + |
| Variant observed in CHST3 (NM_004273.5) | |||||||||
| Zygosity | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | ||||
| Nucleotide change | c.500A > G | c.688G > A | c.976dupG | c.1165G > C | c.430G > A | ||||
| Amino acid change | p.(His167Arg) | p.(Glu230Lys) | p.(Asp326GlyfsTer186) | p.(Ala389Pro) | p.(Gly144Ser) | ||||
| Location | Exon 3 | Exon 3 | Exon 3 | Exon 3 | Exon-3 | ||||
| Variant status | Novel | Reported | Novel | Novel | Novel | ||||
| Reference | Unger et al., 2010 (same amino acid change was reported) | NA | NA | NA | NA | ||||
| ACMG classification | Likely pathogenic (PM5 + PM2 + PP3 + PP1) |
Likely pathogenic (PM1 + PM2 + PP3 + PP4) |
Pathogenic (PVS1 + PM2 + PP3) |
Likely pathogenic (PM1 + PM2 + PP1 + PP3+ PP4) |
Likely pathogenic (PM1 + PM2 + PM5 + PP1 + PP3 + PP4) |
||||
- Note: (+): Present; (−): Absent; NA, Not available.
3.2 Molecular findings
We identified four missense and one frameshift variations in CHST3 (NM_004273.5) gene in the homozygous state in five families. We found three novel variants c.976dupG; p.(Asp326GlyfsTer186), c.1165G > C;p.(Ala389Pro) and c.430 G > A;p.(Gly144Ser) and one known variant c.688G > A;p.(Glu230Lys) in CHST3 gene by Sanger sequencing in four affected families. ES identified a novel variant c.500A > G;p.(His167Arg) in family 1. All variants are located on sulfotransferase domain of protein which begins at 132 aa position to 452 aa position. Four biallelic pathogenic variants were missense and were classified as likely pathogenic as per ACMG 2015 criteria (Richards et al., 2015). The frameshift variant, p.(Asp326GlyfsTer186) might lead to premature termination of protein and be classified as pathogenic. Two variants c.500A > G; p.(His167Arg) and c.976dupG; p.(Asp326GlyfsTer186) are not observed in gnomAD population database while three of them (c.688G > A; p.(Glu230Lys), c.430 G > A, p.Gly144Ser and c.1165G > C, p.Ala389Pro) are found in gnomAD v2.1.1 in heterozygous state in one individual with the allele frequencies 0.000004236, 0.000006432 and 0.000004246 respectively. However, none of the variants were observed in our in-house dataset of 2632 exomes (Kausthubham et al., 2021). The variant details are listed in Table 1 and multiple in silico tools for predicting pathogenicity for these variants are mentioned in Table S1. The new variants were submitted to ClinVar database.
4 DISCUSSION
CHST3-related CDCJD is characterized by congenital joint dislocation/contractures, progressive vertebral changes, metacarpal shortening, and recessive inheritance pattern. CHST3-related CDCJD was earlier known as Omani-type spondyloepiphyseal dysplasia, humerospinal dysostosis, and recessive Larsen syndrome (Mortier et al., 2019). These disorders were previously described as distinct entities until the molecular etiology was identified (Mégarbané & Ghanem, 2004; Rajab et al., 2004; van Roij et al., 2008).
All affected individuals presented in this study have characteristic features for CHST3-related CDCJD. They had congenital joint (knee and radial heads) dislocations, short trunk short stature, and progressive vertebral anomalies. In addition to these manifestations, most of the patients presented with elbow contractures, pectus carinatum, irregular distal femoral epiphysis, and club feet. Other less frequent findings such as camptodactyly, brachydactyly, short femoral neck, and pes planus could also be noted (Duz & Topak, 2020). Moreover, extra skeletal findings such as cardiac involvement were present in four patients. None of the patients had hearing loss. Seven patients reported in this study had supernumerary carpal ossification centers which were observed in only six previously reported patients and most of them were above 10 years of age (Albuz et al., 2020; Otaify et al., 2023; Tuysuz et al., 2009; van Roij et al., 2008). The other two affected individuals were of ages 3 and 6 years, suggesting the possibility of observing this feature later in life. Moreover, severe progressive kyphoscoliosis to the extent of postural problems, could be observed in three of our patients similar to those was described by Rajab et al. and Tuysuz et al. in their study (Rajab et al., 2004; Tuysuz et al., 2009). The widening of interpedicular distance was also more pronounced in these patients.
This report provides further evidence for the cardiac involvement in this condition. First reported by Kozlowski et al. (1974), cardiac defects including hypertrophy of both atria and ventricle, thickening of mitral valve leaflet, aortic regurgitation, and stenosis were noted in several reports (Hall, 1997; Otaify et al., 2023; Rajab et al., 2004; Srivastava et al., 2017; Tuysuz et al., 2009; Unger et al., 2010; Yan et al., 2017). It appears that mitral, tricuspid, and aortic valves are commonly affected in that order with thickened leaflets, stenosis, and insufficiency. In recent study by Otaify et al., severe cardiac valvular insufficiencies were reported in three individuals (Otaify et al., 2023). Considering this observation, a cardiac evaluation is warranted in patients with CHST3 deficiency. Being cognizant about this manifestation could help in early detection of cardiac anomalies and benefit the patient in providing appropriate care.
There are 52 disease-causing variants identified till date, including our four novel variants (Otaify et al., 2023). Most of the pathogenic variations are substitutions (Unger et al., 2010). However, a few small deletions, insertions, and indel are also reported. Most of the known pathogenic variants are present in exon-3 which encodes for sulfotransferase domain in CHST3 protein and conserved among different vertebrates (Figure S2). All the variants mentioned in this study are located in sulfotransferase domain predicted to affect the enzymatic activity of the CHST3 protein. The variant p.(Asp326GlyfsTer186) identified in this study, is predicted to lead to the termination of protein. Amino acid change at position p.(His167Arg) was mentioned in Unger et al study but the nucleotide change c.500A > G resulting this amino acid change is novel. Three individuals with variant p.(Gly144Ser) had severe kyphosis which was resembling to patient with amino acid change p.Thr141Met mentioned in study by Tuysuz et al. (2009).
Five Indian individuals affected with CHST3-related CDCJD were described in literature (Srivastava et al., 2017; Unger et al., 2010). In the study of Unger et al., two patients from two unrelated Indian families harbored the same homozygous missense variant (c.661C > T; p.(Arg221Cys) and in Srivastava et al. study, three affected individuals were reported with two missense variants c.904G > C; p.(Asp302His), c.491C > T; p.(Pro164Leu) and one frame shift insertion c.533_534ins G; p.(Ala179Argfs) in homozygous state. Patient 1 from Srivastava et al study presented with rare clinical findings such as bony outgrowth near left superior iliac spine, hypoplastic patella, and loss of intercondylar notch in distal epiphyses of femora. However, we did not encounter such phenotypic variability in our cohort nor could establish any genotype–phenotype correlation among affected Indian individuals.
The variant c.500A > G was found in homozygous state in non-consanguineous Indian parents in family 1. Finding homozygous variants in families with non-consanguinity is a frequent occurrence in Indian population as endogamy is a common practice (Phadke et al., 2021). In a previous study from our center on skeletal dysplasia in 508 families, we noted homozygous variants in 43.3% of families in whom consanguinity was not reported (Uttarilli et al., 2019).
The phenotype of seven affected individuals (family 2, 3, 4, and 5) in our cohort was consistent with CHST3-related CDCJD and we performed single gene testing. Bifid distal humerus is the characteristic feature of CDCJD and presence of a bifid humerus could act as a phenotypic handle for this disorder. Atelosteogenesis type II, a lethal skeletal dysplasia is also noted with this significant feature. However, the individuals affected with atelosteogenesis type II will be stillborn or succumb shortly after birth. Hence, we believe single gene testing for CHST3 gene with thorough clinical evaluation could be cost-effective and rapid genetic testing modality, especially in Indian scenario.
In conclusion, we describe the phenotypic and genotypic evaluation of nine affected individuals with CHST3-related CDCJD. This is the largest cohort of individuals affected with CHST3-related CDCJD from India till date. This study also adds four novel variants to the CHST3 genotypic spectrum.
Web Resources
PRIMER 3 v.4.1.0, http://primer3.ut.ee/
Ensembl, https://asia.ensembl.org/index.html
NCBI, https://www.ncbi.nlm.nih.gov/
Mutation Taster, http://www.mutationtaster.org/
OMIM, https://www.omim.org/
gnomAD, https://gnomad.broadinstitute.org/
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
HGMD, http://www.hgmd.cf.ac.uk/ac/search.php
Varsome, https://varsome.com/
Mutalyzer, https://mutalyzer.nl/
GATK, https://gatk.broadinstitute.org/
AUTHOR CONTRIBUTIONS
Swati Singh: Writing—original draft, investigation, data curation, writing—review & editing; Prince Jacob, Siddaramappa J. Patil, Mamta Muranjan, and Hitesh Shah: Data acquisition, validation, investigation, writing—review & editing; Katta Mohan Girisha: Data acquisition, funding acquisition, conceptualization, supervision, writing—review & editing, methodology; Gandham SriLakshmi Bhavani: Conceptualization, methodology, supervision, acquisition, writing—review & editing.
ACKNOWLEDGMENTS
We thank the patients and their families for participating in the study. We are also grateful to the Department of Science and Technology, India, for funding the project titled “Application of autozygosity mapping and exome sequencing to identify genetic basis of disorders of skeletal development” (SB/SO/HS/005/2014) to Dr. Girisha KM and acknowledge the financial support provided by Joint CSIR-UGC NET Junior Research Fellowship awarded by Human Resource Development Group under Council of Scientific and Industrial Research (CSIR), Government of India: 08/028(0002)/2019-EMR-I (to Swati Singh). We thank the “SG10K_Pilot Investigators” for providing the SG10K_Pilot data (EGAD00001005337) The data from the “SG10K_Pilot Study” reported here were obtained from EGA. This manuscript was not prepared in collaboration with the “SG10K_Pilot Study” and does not necessarily reflect the opinions or views of the “SG10K_Pilot Study.
CONFLICT OF INTEREST STATEMENT
We declare that the authors do not have any conflict of interest.
CONSENT TO PARTICIPATE
Written informed consent was obtained from the parents/legal guardian/ individual participants included in the study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




