Diagnostic yield of clinical exome sequencing in adulthood in medical genetics clinics
Funding information: Alberta Health Services; Children's Health; Stollery Children's Hospital Foundation; Health Foundation
Abstract
Clinical exome sequencing (ES) is the most comprehensive genomic test to identify underlying genetic diseases in Canada. We performed this retrospective cohort study to investigate the diagnostic yield of clinical ES in adulthood. Inclusion criteria were: (1) Adult patients ≥18 years old; (2) Patients underwent clinical ES between January 1 and December 31, 2021; (3) Patients were seen in the Department of Medical Genetics. We reviewed patient charts. We applied American College of Medical Genetics and Genomics and the Association for Molecular Pathology variant classification guidelines for interpretation of variants. Non-parametric Fisher's exact statistical test was used. Seventy-seven patients underwent clinical ES. Fourteen different genetic diseases were confirmed in 15 patients: FBXO11, MYH7, MED13L, NSD2, ANKRD11 (n = 2), SHANK3, RHOBTB2, CDKL5, TRIO, TCF4, SCN1, SMAD3, POGZ, and EIF2B3 diseases. The diagnostic yield of clinical ES was 19.5%. Patients with a genetic diagnosis had a significantly higher frequency of neurodevelopmental disorders than those with no genetic diagnosis (p = 0.00339). The diagnostic yield of clinical ES was the highest in patients with seizures (35.7%), and with progressive neurodegenerative diseases (33.3%). Clinical ES is a helpful genomic test to provide genetic diagnoses to the patients who are referred to medical genetic clinics due to suspected genetic diseases in adulthood to end their diagnostic odyssey. Targeted next generation sequencing panels for specific phenotypes may decrease the cost of genomic test in adulthood.
1 INTRODUCTION
There are 4697 single gene disorders and 7260 phenotypes with known molecular basis reported in The Online Mendelian Inheritance in Man (OMIM) catalog (accessed October 7, 2022, https://www.omim.org/statistics/geneMap). More than two-third of those genetic diseases have been discovered in the last 10–15 years since next generation sequencing technologies have been applied for the discovery of novel genetic diseases (Claussnitzer et al., 2020). These are achieved by national and international collaborations and with the establishment of the Centers for Mendelian Genomics supported by the National Human Genome Research Institute (NHGRI) within the United States National Institutes of Health (NIH) which established the Centers for Mendelian Genomics (CMGs), with additional support from the National Heart, Lung, and Blood Institute and, later, the National Eye Institute in 2011 (Baxter et al., 2022).
There have been several clinical research studies reporting the diagnostic yield of exome sequencing (ES) in the pediatric population. However, there are only a small number of studies applying ES in the adult population in individuals suspected of having an underlying genetic disease. Adult ES studies have been reported for different clinical phenotypes such as ataxia, stroke, epilepsy, and intellectual disability in neurology clinics (Bardakjian et al., 2018; da Graça et al., 2022; Guo et al., 2021; Ilinca et al., 2020; Minardi et al., 2020; Pierce et al., 2019; Sabo et al., 2020; Snoeijen-Schouwenaars et al., 2019).
Since 2015, clinical ES has been widely used in the medical genetic clinics for clinical diagnostics in Canada. Clinical ES is the most comprehensive clinical genomic test available in publicly funded health care system in Canada. The application of clinical ES has expanded the phenotypic spectrum of rare genetic diseases in our clinical practice.
Unfortunately, genetic tests are not widely applied in adulthood due to limited resources and knowledge for a possible underlying genetic disease in adulthood. However, it is still important to confirm a genetic diagnosis in adults to provide precision therapies based on their genetic diagnoses. In this study, we investigated the landscape of genetic diseases in adults who were referred to our Department of Medical Genetics for suspected underlying genetic diseases. We investigated the diagnostic yield of clinical ES and studied the phenotypes in adulthood. We think that our study will allow us to disseminate this knowledge to adult specialties for the landscape of genetic diseases in adulthood and advocate for the access to genetic tests in adulthood in our province. Genetic diagnosis is crucial for choosing appropriate precision therapies and providing appropriate genetic counseling in adulthood.
2 MATERIALS AND METHODS
The Research Ethics Office, Health Research Ethics Board, University of Alberta (Study ID: Pro00117378) approved this retrospective cohort study in the Department of Medical Genetics, Faculty of Medicine and Dentistry, University of Alberta. The Northern Alberta Clinical Trials and Research Centre (NACTRC) and Alberta Health Services provided operational and administrative approval (PRJ38569).
Our Department of Medical Genetics receives about 2000 patient referrals per year. The referral reasons include suspected underlying genetic disease to perform genetic tests or abnormal genetic tests for genetic counseling. About 30% of those referrals are for adult patients. To investigate landscape of genetic diseases and diagnostic yield of clinical ES in adulthood, we performed this retrospective cohort study in an academic medical genetic center. Our inclusion criteria were: (1) Adult patients ≥18 years old; (2) Patients who underwent clinical ES between January 1 and December 31, 2021; (3) Patients were seen by one of the nine geneticists in the Department of Medical Genetics for genetic diagnostic investigations. Exclusion criteria included: (1) Patient <18 years old; (2) Patients who did not have clinical ES; (3) Patients had clinical ES before January 1, 2021 or after December 31, 2021. We reviewed patients' charts for clinical features, biochemical investigations, molecular genetic investigations, cardiac assessments, and neuroimaging. We entered all information into an Excel database (Microsoft Corp.).
We divided the study cohort into two groups: Group 1: patients with no genetic diagnoses; Group 2: patients with genetic diagnoses, to compare if any of the clinical or abnormal neuroimaging features would be helpful to guide physicians for ordering clinical ES in their adult patients.
Clinical ES using patients' and parents' DNA samples were performed in clinical molecular genetic laboratories according to their methods (e.g., Blueprint Genetics, Prevention Genetics). We applied American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) variant classification guidelines for interpretation of variants (Richards et al., 2015). We also searched all variants in the Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org/about) for their allele frequency in the general population (Lek et al., 2016). Patients were given the option of receiving secondary findings based on the ACMG actionable genes recommendations (Kalia et al., 2017).
We recorded costs in Canadian dollars at the time of test approval to analyze costs of clinical ES and other genetic tests per patient in our center.
Non-parametric Fisher's exact statistical test was used for between-group comparisons. All analyses were performed using R (v.4.0.2) statistical software. A p-value <0.05 was considered statistically significant.
3 RESULTS
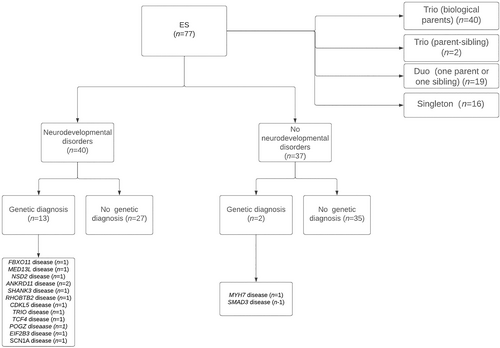
There were 77 patients who underwent clinical ES fulfilling inclusion criteria including 34 males and 43 females. The average current age was 33.22 ± 13.22 years standard deviation (SD) (range 18–72 years). Forty patients had clinical ES trio (both biological parents), 2 patients had mother and an affected sibling clinical ES trio-, 19 patients had -clinical ES duo (18 patients with patient and one parent and one patient with patient and sibling), and 16 patients had clinical ES singleton (biological parents not available). Number of patients and their diagnosis are depicted in Figure 1. Fourteen different genetic diseases were confirmed in 15 patients by the application of clinical ES. Demographics, clinical features, neuroimaging features and genetic diagnoses of those 15 patients are summarized in Table 1. Previous genetic investigations included chromosomal microarray (n = 40) (51.9%), karyotype, Fragile-X test, Prader-Willi and Angelman syndrome test, Di-George syndrome test, and mitochondrial DNA sequencing test which did not confirm any genetic diagnosis.
| Number/study ID/diagnosis/sex/age/consanguinity | Clinical features (age of onset) | Neuroimaging | Other abnormal investigations | Exome sequencing result |
|---|---|---|---|---|
| 1/WESAdult004/FBXO11 disease/male/37/no | ID, weight gain problems, IDDM (3 yrs) | Normal CT | Glucose 0.1 mmol/L (ref range 3.3–6.0) | Heterozygous presumed de novo 5.8 kb deletion (c.2006 + 1_2007-1) (1*_?)del) in FBXO11 (not in mother; father not available) |
| 2/WESAdult011/MYH7 related DCMP/female/26 yrs/no | DCMP, cardiac insufficiency (NA) | NP | Dilated left ventricle, and atrium in echocardiography | Heterozygous c.2168G>A (p.Arg723His) in MYH7 paternally inherited (mother not available) |
| 3/WESAdult013/MED13L disease/female/22 yrs/no | GDD, hypotonia, hyperopia, strabismus, astigmatism, dysarthria, dyspraxia (1 yr) | NP | Marginally low total and free carnitine, likely nutritional | Heterozygous de novo c.6355C > T (pGln2119*) in MED13L |
| 4/WESAdult014/Wolf-Hirschhorn syndrome/male/20 yrs/no | GDD, anxiety disorder, FTT (4 mo) | NP | None | Heterozygous de novo c.2438G.A (p.Cys813Tyr) in NSD2 |
| 5/WESAdult015/KBG syndrome/female/22 yrs/no | GDD, ADHD, depression, aggressive behavior (1 yr) | NP | None | Heterozygous presumed de novo c.6792dup (p.Ala2265Argfs*8) in ANKRD11 (not in mother; father not available) |
| 6/WESAdult016/Phelan-McDermid syndrome/female/25 yrs/no | Pervasive developmental disorder, cognitive dysfunction (3 yrs) | NP | None | Heterozygous de novo c.5148dup (p.Gly1717Argfs*39) in SHANK3 |
| 7/WESAdult019/RHOBTB2 disease/male/36 yrs/no | Lactic acidemia, CP, nonverbal, epilepsy, developmental regression (25 yrs), dystonia, FTT (3 mo) | Increased WM signal in cortical gray matter, lentiform nucleus, caudate head, cerebellar atrophy in MRI | Lactate 5.5 (ref range <2.2) CK 2011 (ref range <250) |
Heterozygous presumed de novo c.1532G>A (p.Arg511Gln) in RHOBTB2 (not in mother; father not available) |
| 8/WESAdult023/CDKL5 disease /female/35 yrs/no | GDD, epilepsy, short stature | Increased WM signal in basal ganglia, cerebral atrophy in MRI | Generalized sharp and slow waves in EEG | Heterozygous de novo c.1981A>T (p.Arg611*) in CDKL5 |
| 9/WESAdult031/TRIO disease /male/20 yrs/no | ID, ADHD, anxiety, microcephaly | NP | None | Heterozygous de novo c.4589A>G (p. Lys1530Arg) in TRIO |
| 10/WESAdult034/SCN1A disease/female/24 yrs/no | GDD, epilepsy | Normal CT | None | Heterozygous presumed de novo c.2792G>A (p.Arg931Pro) in SCN1A (not in mother; father not available) |
| 11/WESAdult048/Pitt-Hopkins syndrome/female/29 yrs/no | GDD, dysmorphic features,a myopia, happy demeanor | NP | None | Heterozygous de novo c.1097G>T (p.Gly366Val) in TCF4 |
| 12/WESAdult055/KBG syndrome/female/29 yrs/no | Microcephaly, short stature, cognitive dysfunction, dysmorphic features,b hearing loss (11 yrs) | NP | None | Heterozygous presumed de novo c.4893del (p.Lys1632Argfs*54) in ANKRD11 (singleton, parents not available) |
| 13/WESAdult059/Loeys-Dietz syndrome/female/47 yrs/no | Aortic dissection type B, tall stature, Hashimoto's thyroiditis, irits, uveitis (36 yrs), hyperflexible joints, myopia, dysmorphic featuresc | Decreased T2 signal in occipital cortex and subcortical WM in MRI | Heterozygous de novo c.714C>G (p.Tyr238*) in SMAD3 | |
| 14/WESAdult077/White-Sutton syndrome/female/21 yrs/no | GDD, ID, FTT, strabismus, dysmorphic featuresd | NP | None | Heterozygous presumed de novo c.2251C>T (p.Gln751*) in POGZ (not in mother; father not available) |
| 15/WESAdult080/EIF2B3 disease/Female/42 yrs/yes (parents first cousins) | Epilepsy, neurodegenerative disease (23 yrs), ID, spasticity, hypothyroidism, dysphagia, acute encephalopathy episodes | Diffuse cerebral, cerebellar, brain stem tracts WM leukoencephalopathy, partial cystic degeneration and WM rarefaction in FLAIR MRI | AS in echo, GGT 222 (ref <50) ALP 679 (ref 40-120), diffuse generalized slowing in EEG |
Homozygous c.260C>T (p.Ala87Val) in EIF2B3 (mother heterozygous; father not available, parents first cousins) |
- Abbreviations: ADHD, attention deficit hyperactivity disorder; AS, aortic stenosis; CAKUT, congenital anomalies of kidney and urinary tract; CK, creatine kinase; CP, cerebral palsy; DCMP, dilated cardiomyopathy; EEG, electroencephalography; FTT, failure to thrive; GDD, global developmental delay; ID, intellectual disability; mo, months; MRI, magnetic resonance imaging; NA, not available; NP, not performed; WM, white matter; yr(s), year(s).
- a Hypertelorism, horizontal palpebral fissures, depressed nasal bridge, thick alea nasi, short stature, arched eyebrows, full lips, prognathism, low set and posteriorly rotated ears, hands with partial cutaneous syndactyly, long big halluces, small feet, short toes, overlapping toes).
- b Low anterior hairline, triangular face, asymmetric face, prominent cheekbones, straight eyebrows, synoprhys, prominent nose, anteverted nares, low hanging columella, large upper central incisors, overfolded helices, hands with short fingers especially 5th finger bilaterally. HC <2%ile, height 5′1″, weight 99 lbs.
- c Split uvula, height 5′11.5″, flat zygomatic arches, hypertelorism, upslanted palbebral fissures, high palate, narrow hands and feet with long fingers and toes.
- d Facial asymmetry, high anterior hair line, ptosis (L > R), upturned nose with blunt tip, full cheeks, long mouth, narrow palpebral fissures, thick upper lip vermillion border, blunt upturned and blunt nasal tip.
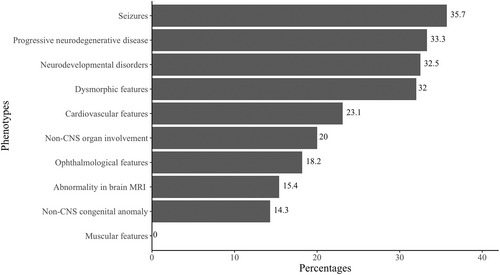
About half of the patients had neurodevelopmental disorders including developmental delay, cognitive dysfunction, intellectual disability, learning difficulties, autism spectrum disorder, attention deficit hyperactivity (ADHD), behavioral disorders (one or more than one of these clinical features). Seizures, congenital anomalies outside of the central nervous system, or cardiovascular features were reported in <20% of the patients. Muscular, ophthalmological, or dysmorphic features were reported in about one-third of the patients (Table 2).
| Total (n = 77) | Group 1: No genetic diagnosis (n = 62) | Group 2: Genetic diagnosis (n = 15) | |||||
|---|---|---|---|---|---|---|---|
| Demographics | Number | Number | (%) | Number | Number | (%) | Statistics (Fisher's exact test) |
| Male sex | 34 | 30 | (48.39) | 4 | 30 | (48.39) | 0.243 |
| Parental consanguinity | 6 | 5 | (8.06) | 1 | 5 | (8.06) | 1 |
| Different clinical features and number of patients for each (%) | |||||||
| Neurodevelopmental disordersa | 40 | (51.95) | 27 | (43.55) | 13 | (86.7) | 3.39 × 10−3* |
| Seizures | 14 | (18.9) | 9 | (14.52) | 5 | (33.3) | 0.1316 |
| Non-CNS congenital anomaly | 14 | (18.2) | 12 | (19.35) | 2 | (13.3) | 0.7247 |
| Dysmorphic features | 25 | (32.5) | 17 | (27.42) | 8 | (53.3) | 0.0694 |
| Cardiovascular features | 13 | (16.9) | 10 | (16.13) | 3 | (20) | 0.7096 |
| Ophthalmological features | 22 | (28.6) | 18 | (29.03) | 4 | (26.7) | 1 |
| Muscular featuresb | 24 | (31.2) | 24 | (38.7) | 0 | (0) | 0.00359* |
| Progressive neurodegenerative disease | 3 | (3.9) | 2 | (3.23) | 1 | (6.7) | 0.4830 |
| Non-CNS organ involvementc | 15 | (19.5) | 12 | (19.35) | 3 | (20) | 1 |
| Abnormality in brain MRI (n = 45) | 26 | (57.8) | 22 (n = 38) | (57.9) | 4 (n = 7) | (57.1) | 1 |
- a Includes developmental delays, cognitive dysfunction, intellectual disability, learning difficulties, autism spectrum disorder, ADHD, and behavioral disorders.
- b Includes muscle twitches, fatigue, stiffness, and ptosis. Four patients had muscle biopsy (1) normal muscle biopsy; (2) complex IV deficiency and subsarcolemmal accumulations of the mitochondria in electron microscopy; (3) ragged red fiber in Gomori trichrome staining and subsarcolemmal accumulation of mitochondria in electron microscopy; (4) type 1 fiber hypotrophy.
- c Includes diabetes, gastrointestinal concerns, renal concerns, hearing loss, liver concerns, connective tissue disorder, pancreatic concerns.
Fourteen different rare genetic diseases were confirmed in 15 adult patients including FBXO11 (OMIM#618089), MYH7 (OMIM#613426), MED13L (OMIM#616789), NSD2 (OMIM#619695), ANKRD11 (OMIM#148050) (KBG syndrome) (n = 2), SHANK3 (OMIM#606232) (Phelan-McDermid syndrome), RHOBTB2 (OMIM#618004), CDKL5 (OMIM#300672), TRIO (OMIM#609823), TCF4 (OMIM#610954) (Pitt-Hopkins syndrome), SCN1A (OMIM#619317), SMAD3 (OMIM#613795) (Loeys-Dietz syndrome), POGZ (OMIM#616364) (White-Sutton syndrome), and EIF2B3 (OMIM#603896) (leukoencephalopathy with vanishing white matter) diseases. The diagnostic yield of trio clinical ES was 17.5% (7 out of 40 patients). The diagnostic yield of duo (child and one parent) clinical ES was 36.8% (7 out of 19 patients). The diagnostic yield of singleton clinical ES was 6.3% (1 out of 16 patients).
There were 86 variants in 77 patients. All variants and their ACMG/AMP classification are summarized in Table S1 (variants in previously established disease genes), Table S2 (variants in genes for return of secondary findings), Table S3 (variants in genes identified as incidental findings), and Table S4 (variants in candidate genes not previously associated with disease). There were 17 pathogenic or likely pathogenic variants in 16 patients and phenotypes were associated with those pathogenic or likely pathogenic variants in 15 of those patients confirming a genetic diagnosis (Table S1). Fourteen patients had autosomal dominant (de novo n = 7; presumed de novo n = 6; inherited n = 1) and one patient had autosomal recessive genetic diseases. Thirty-eight VUSs were reported in 24 patients and their results were inconclusive (Table S1). The number of VUSs reported in patients who underwent clinical ES trio (n = 9) or duo (n = 9) were similar.
Seventy-one out of 77 families (92% of the study cohort) consented for secondary findings. Seven pathogenic or likely pathogenic variants were reported in genes for return of secondary findings (Table S2). The diagnostic yield of secondary findings was 9.9% in the study cohort. All individuals with abnormal secondary findings were provided genetic counseling and referred to appropriate clinics for their management. Eight pathogenic or likely pathogenic variants in seven different genes were reported in nine patients as incidental findings (Table S3). Fourteen variants of unknown significance in 12 different candidate genes in 11 patients are summarized in Table S5. None of those genes were well characterized at the time of clinical ES to confirm a genetic diagnosis. All patients with inconclusive or negative clinical ES were recommended to be re-referred in 2–5 years for VUS re-classification and clinical ES re-analysis.
Group 1 had a significantly higher frequency of muscular features compared to Group 2 (p = 0.00359). Group 2 had a significantly higher frequency of neurodevelopmental disorders compared to Group 1 (p = 0.00339) (Table 2). The diagnostic yield of clinical ES for different phenotypes is depicted in Figure 2. The diagnostic yield of clinical ES was 32.5% in patients with neurodevelopmental disorders, whereas it was 5.4% in patients without neurodevelopmental disorders which was statistically significant (p = 0.003386).
The estimated total cost of clinical ES was $203,000 Canadian dollars for the study cohort and the average cost of clinical ES was $2655 Canadian dollars per patient. The estimated total cost of other genetic investigations was $83,880 Canadian dollars for the study cohort and the average cost of other genetic tests was $1089.4 Canadian dollars per patient. The median number of other genetic tests was 1 ± 1.3 SD (range 0–6 genetic tests per patient). There were also different types of non-genetic investigations that we did not perform any cost calculations including chemistry tests, biochemical investigations, biopsies, and neuroimaging.
4 DISCUSSION
We report a 19.5% diagnostic yield of clinical ES in adult patients who were referred for the investigations of underlying genetic causes for their phenotypes to the Department of Medical Genetics at our institution during the study period. Patients with genetic diagnoses had a significantly higher frequency of neurodevelopmental disorders (p = 0.00339). Patients with no genetic diagnoses had a significantly higher frequency of muscular features (p = 0.00359). The diagnostic yield of clinical ES was significantly higher in patients with neurodevelopmental disorders (p = 0.003386). The diagnostic yield of duo clinical ES was the highest in our study cohort. There was a statistically significant higher diagnostic yield of duo clinical ES compared to singleton clinical ES (p = 0.04725*), whereas there was no statistically significant difference between trio and duo clinical ES (p = 0.1167) and between trio and singleton ES (p = 0.4163). The diagnostic yield of clinical ES was more than 30% of patients with different clinical features (listed in Table 2) such as seizures, dysmorphic features, neurodevelopmental disorders, and progressive neurodegenerative disorders in our study. Clinical ES is helpful to provide genetic diagnoses in undiagnosed adult patients to end their diagnostic odyssey. If routine genetic tests, such as chromosomal microarray, Fragile-X, or other genetic tests, based on the phenotypes, did not confirm a genetic diagnosis, we recommend a stepwise approach for the appropriate molecular genomic test in adults for genetic diagnosis based on the phenotypes (Figure S1). We think that application of this algorithm may decrease the cost of molecular genomic tests and increase the diagnostic yield of clinical ES in the future.
Indeed, in our study six of our patients would have been diagnosed by one of the targeted next generation sequencing panels including (1) a comprehensive epilepsy panel (n = 4) in the diagnosis of CDKL5, SCN1A, RHOBTB2, and EIF2B3 diseases; (2) a connective tissue disease panel (n = 1) in the diagnosis of SMAD3 disease; (3) a leukodystrophy and leukoencephalopathy panel (n = 1) in the diagnosis of EIF2B3 disease; and (4) a cardiomyopathy panel (n = 1) in the diagnosis of MYH7 associated dilated cardiomyopathy. The targeted next generation sequencing panels based on patients' phenotypes would have decreased the cost of genetic investigations about 50% in these six patients.
There are a small number of studies using ES (research or clinical) in adults which are summarized in Table S5. The diagnostic yield ranged from 17% to 60% with a wide range of phenotypes in those studies (da Graça et al., 2022; Guo et al., 2021; Minardi et al., 2020; Mu et al., 2019; Posey et al., 2016; Sabo et al., 2020; Shickh et al., 2021). Neurogenetic phenotypes such as intellectual disability (Sabo et al., 2020), epileptic encephalopathy (Minardi et al., 2020), ataxia (da Graça et al., 2022), and neuromuscular disorders (Guo et al., 2021) reach a higher diagnostic yield of ES. Ophthalmological features seem to have a lower diagnostic yield of ES (2.1%) in a previous study (Guo et al., 2021) whereas we report an 18.2% diagnostic yield of clinical ES in patients with ophthalmological features in our study. The diagnostic yield of ES seems lower when there are large number of patients in a study cohort and when the phenotypes are not well-defined (17.5%) (Posey et al., 2016). We think that our study results represent real-world yield of clinical ES as medical genetic clinics receive referrals with a wide range of phenotypes for genetic investigations. To the best of our knowledge, this is the first clinical study reporting diagnostic yield of clinical ES from a single medical genetics department and all clinical ES was requested by medical geneticists in our study.
Our study limitations include (1) There were nine different physician clinical practices for clinical ES indications in our study; (2) We have small number of patients in our study cohort; (3) We do not know if a longer study period would have increased the number of genetic diagnoses and the diagnostic yield of clinical ES. Despite these limitations, we think that our study is valuable since it increases our knowledge regarding the diagnostic yield of clinical ES, while reporting on the landscape of genetic diseases in adulthood.
In summary, we report a 19.5% of diagnostic yield of clinical ES in adults. Existing targeted next generation sequencing panels for specific phenotypes may decrease the cost of molecular genomic tests and increase the diagnostic yield of clinical ES in adults in the future. A follow-up study using our stepwise approach for the appropriate molecular genomic tests in adults may be helpful to investigate its cost effectiveness and diagnostic yield in the future. This may give us an opportunity to further develop and validate this algorithm for wider use. We think that it is important to confirm genetic diagnoses in adults to provide them with appropriate genetic counseling while supporting their medical management even their diagnostic odyssey is a long journey.
AUTHOR CONTRIBUTIONS
Conceptualization, Project Administration, Supervision, Validation: Saadet Mercimek-Andrews. Data Curation, Investigation, Writing, review and editing: Apurba Mainali; Taryn Athey; Shalini Bahl; Clara Hung; Oana Caluseriu; Alicia Chan; Alison Eaton; Shailly Jain Ghai; Peter Kannu; Melissa MacPherson; Karen K. Niederhoffer; Komudi Siriwardena; Saadet Mercimek-Andrews. Formal Analysis: Apurba Mainali; Saadet Mercimek-Andrews. Methodology: Apurba Mainali; Taryn Athey; Shalini Bahl; Saadet Mercimek-Andrews. Writing original draft: Apurba Mainali; Taryn Athey; Shalini Bahl; Saadet Mercimek-Andrews.
ACKNOWLEDGMENTS
We would like to thank all genetic counselors, genetic assistants, and administrative assistants for their excellent clinical care. We would like to thank Edmonton Zone Alberta Health Services and NACTRC for their support for us to perform this clinical research study. We would like to thank the Genetic Resources Centre and Alberta Precision Laboratories for clinical ES. We would like to thank Mrs. Farah Hassan, research coordinator for managing the study setup, including ethics submissions and operational approval requests. We would like to thank the Women and Children's Health Research Institute for providing research support services to help manage this research project. These services are available because of the generosity of the Stollery Children's Hospital Foundation and the Alberta Women's Health Foundation.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.




