Phenotype delineation of ZNF462 related syndrome
Funding information CIHR Foundation Grant, Grant/Award Number: FDN-154279; National Eye Institute, Grant/Award Number: R01EY027421; National Heart, Lung, and Blood Institute, Grant/Award Number: X01HL132377; National Human Genome Research Institute, Grant/Award Number: Intramural Research Program
Abstract
Zinc finger protein 462 (ZNF462) is a relatively newly discovered vertebrate specific protein with known critical roles in embryonic development in animal models. Two case reports and a case series study have described the phenotype of 10 individuals with ZNF462 loss of function variants. Herein, we present 14 new individuals with loss of function variants to the previous studies to delineate the syndrome of loss of function in ZNF462. Collectively, these 24 individuals present with recurring phenotypes that define a multiple congenital anomaly syndrome. Most have some form of developmental delay (79%) and a minority has autism spectrum disorder (33%). Characteristic facial features include ptosis (83%), down slanting palpebral fissures (58%), exaggerated Cupid's bow/wide philtrum (54%), and arched eyebrows (50%). Metopic ridging or craniosynostosis was found in a third of study participants and feeding problems in half. Other phenotype characteristics include dysgenesis of the corpus callosum in 25% of individuals, hypotonia in half, and structural heart defects in 21%. Using facial analysis technology, a computer algorithm applying deep learning was able to accurately differentiate individuals with ZNF462 loss of function variants from individuals with Noonan syndrome and healthy controls. In summary, we describe a multiple congenital anomaly syndrome associated with haploinsufficiency of ZNF462 that has distinct clinical characteristics and facial features.
1 INTRODUCTION
Heterozygous loss of function variants in ZNF462 present with a recognizable pattern of phenotype characteristics (Weiss et al., 2017). The first reported case was a reciprocal translocation t(2;9)(p24;q32) that disrupted both ZNF462 and ASXL2 (Ramocki et al., 2003; Talisetti et al., 2003). This individual presented with ptosis, agenesis of the corpus callosum, ventricular septal defect, periventricular nodular heterotopia, retina and iris colobomas, and a dysplastic left ear and hearing loss. ASXL2 was subsequently associated with Shashi-Pena syndrome which presents as macrocephaly, retrognathia, low set ears, hypertelorism, arched eyebrows, intellectual disability, scoliosis, congenital heart disease, and hypotonia (Shashi et al., 2016). The phenotype of the individual in this case report likely resulted from the loss of function of both ZNF462 and ASXL2. Over a decade later, Weiss et al. described six individuals from four families with putative loss of function variants and two unrelated individuals with deletions involving adjacent genes (Weiss et al., 2017). The individuals described by Weiss et al. presented with ptosis (100%), trigonocephaly or metopic ridging (83%), and developmental delay or autism spectrum disorder (33%) (Weiss et al., 2017). Subsequently, Cosemans et al. described an individual with a de novo translocation that disrupted ZNF462 and KLF12 who presented with clinical features similar to those described by Weiss et al. (2017) and Cosemans et al. (2018).
Zinc finger protein 462 (ZNF462) is a C2H2 type zinc finger transcription factor of unknown function (Nagase, Nakayama, Nakajima, Kikuno, & Ohara, 2001). Although the specific function of this molecule is unknown, animal studies have shown that it plays a vital role in embryonic development. In Xenopus laevis, knockdown expression of Zfp462 disturbs early embryonic development and results in altered cell division during the cleavage stage; this phenotype is rescued with human ZNF462 mRNA (Laurent et al., 2009). In the mouse model, Zfp462 knockout (KO) mice were prenatal lethal and heterozygous KO mice (Zfp462+/−) had developmental delay, low body and brain weights, and anxiety-like behaviors with excessive self-grooming behavior (Wang et al., 2017).
In this report, we describe 14 new individuals in addition to the 10 previously reported cases in the medical literature with truncating variants in ZNF462, collectively review the clinical presentation of this syndrome, and test facial analysis technology's ability to diagnose this syndrome.
2 METHODS
2.1 Clinical
The study was approved by National Human Genome Research Institute Institutional Review Board. Thirteen new individuals in this report with loss of function variants in ZNF462 were diagnosed using whole exome sequencing (WES) in multiple research and commercial labs including GeneDx and Ambry, and one individual (Patient 5) was diagnosed by whole genome sequencing. Nine of the 14 individuals were ascertained through GeneMatcher (Sobreira, Schiettecatte, Valle, & Hamosh, 2015).
2.2 Facial analysis technology
We performed two binary classification experiments using the Face2Gene Research application (FDNA Inc., Boston, MA), as previously described (Gurovich et al., 2019). Frontal facial 2D images were collected for three cohorts: individuals with ZNF462 loss of function variants, Noonan syndrome, and healthy controls. Noonan syndrome was used as a second control group due the overlapping facial features of ptosis, downslanting palpebral fissures, hypertelorism, and low set ears in a subset of individuals. All facial images were fully de-identified through the use of the DeepGestalt facial analysis (Gurovich et al., 2019). Controls were matched for age, gender, and ethnicity.
3 RESULTS
3.1 Clinical
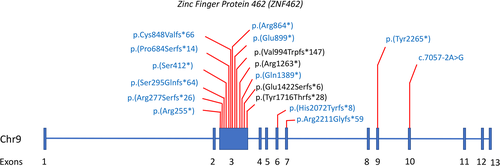
Figure 1 shows the single nucleotide variant and small insertion/deletion (indel) locations on ZNF462 for both the 14 newly reported cases and previously reported cases. All variants are predicted to result in loss of function, including a canonical splice variant in Patient 6 that is predicted to result in abnormal splicing (Table 1; Figure 1). Most of these variants are in Exon 3, which makes up 54% of the coding region of ZNF42.
| Patients | Age | Sex | ZNF462 variant (NM_021224.5) | Inheritance | DD | ASD | Ptosis | Down slanting palpebral fissures | Arched eyebrows | Short upturned nose with bulbous tip | Exaggerated cupid bow/wide philtrum | Feeding issues | Epicanthal folds | Ears | Craniosynostosis/metopic ridging | Hypotonia | Hypertelorism | Corpus callosum dysgenesis | CHD | Limb anomalies (minor) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 16 months | M | c.2590C>T p.(Arg864*) | Maternal (mosaic) | Motor/speech | − | + | − | + | − | + | + | + | Low set | − | + | + | Normal MRI | Not reported | Fifth finger clinodactyly |
| 2 | 10 years | M | c.2542del p.(Cys848Valfs*66) | de novo | Motor/speech | + | + | + | + | + | + | + | + | − | − | − | + | Normal MRI | Not reported | Not reported |
| 3 | 6 years | M | c.831_834del p.(Arg277Serfs*26) | de novo | Motor/speech | − | − | − | − | + | − | + | − | Inner ear malformation | − | + | + | Normal MRI | Bicuspid aortic valve; VSD | Not reported |
| 4 | 2 years 7 months | M | c.6214_6215del p.(His2072Tyrfs*8) | de novo | Speech | − | + | − | − | − | + | + | + | Small, lowset | + | − | − | Not tested | Not reported | Not reported |
| 5 | 14 years | F | c.763C > T p.(Arg255*) | Paternal | IEP/special education | − | + | − | + | − | − | − | + | Hearing loss | + | − | − | Not tested | Not reported | Not reported |
| 6 | 7 months | F | c.7057-2A > G | de novo | Early intervention for DD | − | + | + | + | + | + | + | + | Horizontal crus helix | − | + | + | Normal MRI | VSD | Prominent creases on hands and feet |
| 7 | 13 years | M | c.6794dup p.(Tyr2265*) | de novo | Cognitive impairment | + | − | − | + | − | + | − | − | Prominent ears/ear pits/hearing loss | + | − | − | Not tested | Not reported | Not reported |
| 8 | 2 years | M | c.882dup p.(Ser295Glnfs*64) | de novo | Speech delay | − | + | + | − | − | − | − | − | − | − | − | − | ACC | Not reported | Not reported |
| 9 | 15 years | M | c.4165C > T p.(Gln1389*) | de novo | Global | − | + | + | + | − | + | + | − | Lowset | − | + | − | Not tested | Not reported | Not reported |
| 10 | 8 years | M | c.1234_1235insAA; p.(Ser412*) | Unknown | Speech delay; motor apraxia; IEP | − | + | − | − | − | − | + | Mildly cupped ears | − | − | − | Normal MRI | Not reported | Not reported | |
| 11 | 2 years 5 months | F | c.6214_6215del p.(His2072Tyrfs*8) | de novo | − | − | + | − | − | − | − | + | − | + | − | Not tested | Not reported | Not reported | ||
| 12 | 9 months | M | c.2049dup p.(Pro684Serfs*14) | de novo | Motor | − | + | + | + | + | + | − | + | − | − | + | + | Normal MRI | Not reported | Not reported |
| 13 | 8 years 7 months | M | c.6631del p.(Arg2211Glyfs*59) | de novo | − | − | + | + | − | + | + | − | − | − | − | − | + | Not tested | Not reported | 5th finger clinodactyly |
| 14 | 8 years | F | c.2695G > T; p.(Glu899*) | mother negative; father unknown | Cognitive impairment | − | − | − | + | − | − | + | − | − | − | + | − | Normal MRI | Not reported | Not reported |
| 15a | 2 years | F | c.3787C > T p.(Arg1263*) | Paternal | − | − | + | + | + | + | + | Not reported | − | − | + | − | + | ACC; colpocephaly | Not reported | Not reported |
| 16a | 4 years | F | c.3787C > T p.(Arg1263*) | Paternal | − | − | + | + | − | + | + | Not reported | + | − | + | − | − | Normal prenatal ultrasound | Not reported | Not reported |
| 17a | 34 years | M | c.3787C > T p.(Arg1263*) | Maternal | − | − | + | − | − | − | − | Not reported | − | − | + | − | − | Not tested | Not reported | Not reported |
| 18a | 2 years | M | c.2979_2980delinsA p.(Val994Trpfs*147) | de novo | Speech | + | + | + | + | + | − | + | + | Left overfolded ear | + | − | − | - normal MRI | Not reported | Single palmar crease;5th finger clinodactyly |
| 19a | 32 months | M | c.4263del p.(Glu1422Serfs*6) | de novo | Motor/speech | + | + | + | − | + | − | − | + | Lowset | + | + | + | Hypoplastic corpus callosum and ventriculomegaly | D-TGA | Not reported |
| 20a | 5 years | F | Chr9:g.(108940763-110561397)del (hg19) | de novo | − | − | + | + | + | + | + | Not reported | + | − | + | − | Hypoplastic corpus callosum | Not tested | Not reported | |
| 21a | 15 years | F | Chr9:g(108464368-110362345)del (hg19) | de novo | Motor/intellectual | + | − | − | − | − | + | Not reported | − | − | − | + | Not tested | VSD | Not reported | |
| 22a | 9 years | M | c.5145delC p.(Tyr1716Thrfs*28) | de novo | Motor/speech delay | + | + | − | − | − | − | Not reported | − | − | − | + | − | Normal MRI | Not reported | Not reported |
| 23b | 5 years | F | t(2;9)(p24;q32); disrupting ZNF462 and ASXL2 | de novo | Intellectual disability | + | + | + | + | + | + | + | − | Lowset; hearing loss | − | + | − | ACC; dilated ventricles | VSD; left ventricular hypertrophy | Single palmar crease; hypoplastic finger nails |
| 24c | 24 years | M | t(9; 13)(q31.2; q22.1) disrupting ZNF462 and KLF12 | de novo | Intellectual disability | + | + | + | − | − | − | + | + | Lowset | + | + | − | Hypoplastic corpus callosum | None reported | Small hands and feet; proximally placed thumbs |
| Cohort prevalenced | 79% | 33% | 83% | 58% | 50% | 46% | 54% | 50% | 46% | 50% | 38% | 50% | 25% | 25% | 21% | 25% | ||||
- Abbreviations: ACC, agenesis of the corpus callosum; ASD, autism spectrum disorder; CHD, congenital heart disease; DD, developmental delay; D-TGA, D-transposition of the great arteries; F, female; IEP, individualized education program; M, male; MRI, magnetic resonance imaging; VSD, ventricular septal defect.
- a Weiss et al. (2017).
- b Talisetti et al. (2003).
- c Cosemans et al. (2018).
- d In order to avoid overestimating phenotype prevalence, we divided each positive phenotype report by the entire cohort (n = 24).
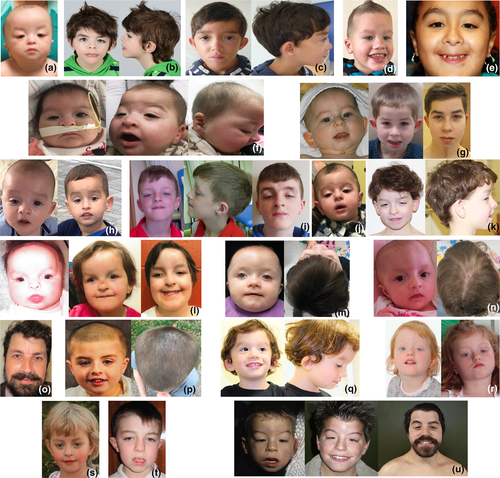
Table 1 summarizes the clinical features of all 24 affected individuals with 96% of individuals being Caucasian. Seventeen of 21 families (86%) have de novo variants, the other four families include unknown, mosaic, and autosomal dominant inheritance (Table 1). The two families with autosomal dominant inheritance demonstrated that the ZNF462 variant segregated with the phenotype characterized in this study: Patient 5's father had ptosis surgery and Patients 15–17 are from the same family, and previously described (Weiss et al., 2017). The one case of mosaicism was in the mother of Patient 1 who had 175 reference reads and 35 alternate reads on WES from a peripheral blood sample (alternate allele frequency = 35/(35 + 175) = 17%), compared to 120 reference reads and 79 alternate reads in the proband (alternate allele frequency = 79/(79 + 120) = 40%). The majority of individuals had developmental delay (79%) and 33% reported autism spectrum disorders (Table 1). The most common facial features were ptosis (83%), down slanting palpebral fissures (58%), exaggerated Cupid's bow/wide philtrum (54%), arched eyebrows (50%), and short upturned nose with bulbous tip (46%). Feeding issues (50%) and hypotonia (50%) were common. Less than half of affected individuals reported metopic ridging or craniosynostosis (38%) or dysgenesis of the corpus callosum (25%). Less common characteristics included structural heart defects (21%) and minor limb anomalies (25%). The clinical analysis of the individuals in this study was heterogenous and not all individuals received brain and heart imaging (Data S1), thus the above fractions may be an underestimation of brain and heart malformations. Figure 2 shows facial images of individuals with loss of function variants in ZNF462.
3.2 Facial analysis technology
Binary comparison between individuals with loss of function variants in ZNF462 and controls was resulted in two statistics: the mean results involved the computation of the average of the AUC of each of the 10 results, and second, the aggregated results consist of a score distribution curve and a receiver-operating-characteristic curve for the aggregated results for each photo used in the validation set. The binary comparison between ZNF462 (n = 21) and healthy controls (n = 21) yielded an area under the curve (AUC) of 0.96 (STD 0.03), demonstrating good separation between these two cohorts (Table S1). Similarly, the comparison between the ZNF462 cohort (n = 21) and the Noonan syndrome cohort (n = 16) yielded an AUC of 0.97 (STD 0.02) which is also good separation (Table S1). The aggregated binary comparison for the ZNF462 group versus health controls yielded an AUC of 0.955 (p = .006) and for the ZNF462 group versus health Noonan syndrome yielded an AUC of 0.972 (p = .001) (Figure S1).
Applying DeepGestalt, the confusion matrix/multi-class comparison of the 58 frontal images of the ZNF462 group and both control groups yielded a mean accuracy of 82.88% (STD 11.79%) which is significantly better than randomly expected (36.21%).
4 DISCUSSION
We report 24 individuals with loss of function variants in ZNF462 which includes 14 previously unpublished individuals and 10 individuals reported in the medical literature. Based on this larger assembled cohort of individuals, the phenotype of loss of function in ZNF462 is now a distinct multiple congenital anomaly syndrome. We show that ptosis (83%), developmental delay (79%), and down slanting palpebral fissures (58%) are three most reported phenotypic features (Table 1). In the previous case series of six families and eight individuals, metopic ridging/craniosynostosis (63%) was a major phenotypic feature. In this report, we show that metopic ridging/craniosynostosis is still important, but less prevalent (25%) in this syndrome. Consistent with the previous report by Weiss et al., 2017 (Table 1: Patients 15–17), loss of function in ZNF462 appears to have variable expressivity and complete penetrance as demonstrated by Patient 5 in the present study with a paternally inherited variant and a father requiring surgery for his ptosis (Table 1; Data S1). Facial analysis technology was able to accurately differentiate individuals with loss of function in ZNF462 from Noonan syndrome and healthy controls. We predict that the widespread use of facial analysis technology will result in an increase in the number of cases diagnosed this syndrome.
Based on the prevalence of developmental delay, corpus callosum anomalies, congenital heart defects, and hearing loss, we recommend a comprehensive multidisciplinary evaluation of individuals with loss of function variants in ZNF462. This evaluation includes at a minimum: a developmental evaluation, a cardiac exam with echocardiography, brain imaging, hearing evaluation, and consultation with a clinical geneticist and genetic counselor. Other evaluations specific to an individual's presentation such as neurosurgery consultation for craniosynostosis may be appropriate. At this time, treatment of complications associated with ZNF462 related syndrome is not different from the general population. As more individuals are studied, future specific management recommendations for ZNF462 related syndrome may be needed.
Pathogenicity of variants in ZNF462 is presumed to be haploinsufficiency based on individuals having loss of function variants only, and this is reinforced by the Genome Aggregation Database (gnomad) constraint metric of observed/expected loss of function (o/e) value (Karczewski et al., 2019). Values less than 0.35 (o/e) are considered under selection against loss of function (LOF) (https://gnomad.broadinstitute.org) and ZNF462 is well below this threshold with an o/e value of 0.03 (90%CI, 0.01–0.09). As noted in the introduction, ZNF462 is important to embryonic development in multiple species. ZNF462 contains 23 C2H2-type zinc finger domains, making DNA binding a likely function (Chang, Stoykova, Chowdhury, & Gruss, 2007). We now know that ZNF462 is involved in chromatin remodeling. Using histone peptide pull down assays in mouse brain and kidney, Eberl et al. showed that ZNF462 binds H3K9 me3, identifying Znf462 as a chromatin reader involved in heterochromatin modification (Eberl, Spruijt, Kelstrup, Vermeulen, & Mann, 2013). Additionally, Eberl et al. report an interaction with Heterochromatin Protein 1α (HP1α) (Eberl et al., 2013). As hallmarks of heterochromatin, HP1α and H3K9me3 are critical for transcriptional silencing of gene and repetitive DNA and for the maintenance of genome integrity (Almouzni & Probst, 2011; Beisel & Paro, 2011; Ren & Martienssen, 2012), further supporting ZNF462's role in chromatin remodeling. Masse et al. used short hairpin RNA knockdown of pluripotent mouse cells, demonstrating a disruption of pericentromeric domains and redistribution of HP1α proteins, giving evidence that Znf462 is instrumental in maintaining heterochromatin in pluripotent cells (Masse et al., 2010).
In summary we present 24 individuals with loss of function variants in ZNF462, and we define a multiple congenital anomaly syndrome that is recognizable from phenotype elements and by using facial analysis technology.
ACKNOWLEDGMENTS
We thank the affected individuals and their families for participating in this study. This project was supported by the National Human Genome Research Institute's Intramural Research Program (P.K. and M.M.); NEI grant R01EY027421 and NHLBI grant X01HL132377 (E.C.E), and a CIHR Foundation Grant (FDN-154279) (K.M.B.). Sequencing and analysis for the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 to Daniel MacArthur and Heidi Rehm. The research conducted at the Murdoch Children's Research Institute was supported by the Victorian Government's Operational Infrastructure Support Program.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.




