p.Arg69Trp in RNASEH2C is a founder variant in three Indian families with Aicardi–Goutières syndrome
Abstract
Aicardi–Goutières syndrome is an early-onset severe neurological disorder characterized by intracranial calcification, white matter abnormalities, hepatosplenomegaly, cerebrospinal fluid lymphocytosis, and elevated interferon-α levels, thus mimicking congenital viral infections. It is a genetically heterogeneous condition and autosomal recessive and autosomal dominant forms with variations in seven genes known till date. Variations in RNASEH2C cause an autosomal recessive form of AGS. Here we report three Indian families with variant, c.205C>T (NM_032193.3, p.Arg69Trp) in RNASEH2C gene identified by whole-exome sequencing and targeted molecular testing of the variant. Review of literature and our data suggest this is likely to be a founder variant in Asians and it would be a good initial variant to screen in patients with Aicardi–Goutières syndrome in Indians.
1 INTRODUCTION
Aicardi–Goutières syndrome (AGS) (MIM #225750) is primarily characterized by early-onset encephalopathy, microcephaly, basal ganglia calcifications, leukodystrophy, hepatosplenomegaly, and thromobocytopenia. Other phenotypic features associated with AGS are chilblains, glaucoma, hypothyroidism, intracerebral calcifications, bowel inflammation, vasculitis, peripheral neuropathy, cardiomyopathy, and systemic lupus erythematosus (Crow et al., 2015). AGS has significant phenotypic overlap with intrauterine viral infections as well as pseudo-TORCH (toxoplasmosis, rubella, cytomegalovirus, herpes simplex, and syphilis) syndrome. Variable clinical expression and severity are known in AGS, although typically it is a severe infantile manifestation. The disorder is genetically heterogeneous disorder and is inherited as an autosomal recessive and less commonly as autosomal dominant trait with pathogenic variations in seven known causative genes (ADAR, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, IFIH1, TREX1) to date (Rice et al., 2014). The recurrent c.205C>T (p.Arg69Trp) variant in RNASEH2C (MIM #610330) has been observed predominantly in South Asian population of Pakistani origin and three patients of Indian origin previously (Crow et al., 2006; Ramantani et al., 2010; Rice et al., 2007; Vogt et al., 2013). Here we report three additional Indian families with the variant c.205C>T (NM_032193.3, p.Arg69Trp) in RNASEH2C gene.
2 MATERIALS AND METHODS
2.1 Clinical report
2.1.1 Patient 1
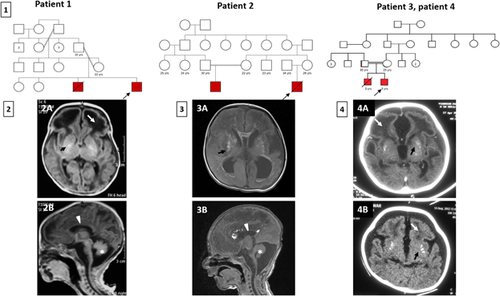
The proband was assessed at 6 months of age for global developmental delay and seizures. One episode of seizure was noted at 3 months of age. He was born to a third degree consanguineously married couple (Figure 1). He was delivered at 40 weeks of gestation by caesarean section, with a birth weight of 2.5 kg. No perinatal or postnatal complications were noted. Feeding was normal. No milestones have been achieved till date. At 6 months, his length was 59 cm (−3 SD), head circumference was 37 cm (−6 SD) and weighed 4.6 kg (−2 SD). Microcephaly, mild coarse facial features were noted. Tone was increased and deep tendon reflexes were brisk. Mild hepatomegaly (4 cm below right costal margin) was observed. Ultrasonography (USG) of the abdomen showed hepatomegaly. Normal fundus was noted on ophthalmology evaluation. Routine hematology examination also returned normal results. Brain imaging at 6 months revealed diffuse white matter disease, hypoplastic corpus callosum, cystic changes in the fronto-temporal white matter and cerebellar hypoplasia, intracranial calcifications (Figure 1(2a and 2b)). Patient expired at the age of 9 months.
2.1.2 Patient 2
He was born to a third degree consanguineous marriage and was seen on postnatal day 18 (Figure 1). Following an uncomplicated pregnancy, he was delivered at full term with a birth weight of 1.8 kg (−1 SD), head circumference 31.5 cm (−1 SD), and length 46 cm (−1 SD). Abnormal movements of all limbs and tonic posturing were observed since birth. At 18 days, he was 48 cm (−1 SD) in length, weighed 2 kg (−1 SD), had a head circumference of 32 cm (−1 SD). Other findings on examination were long fingers, spasticity, brisk deep tendon reflexes, unilateral undescended testis and micropenis. Brain imaging revealed prominent ventricles, prominent cisterna magna, few echogenic spots in the brain parenchyma suggestive of calcifications, hypoplasia of corpus callosum, hypoplastic vermis and cerebellum (Figure 1(3a and 3b)). Electroencephalogram revealed left temporal sharp waves and patent ductus arteriosus was reported on echocardiography. Splenomegaly and altered echotexture of liver was noted on USG. Elevated level of thyroid stimulating hormone (22 mU/L, ref: <6 mU/L) was noted and the baby was started on L-thyroxine. Ophthalmology evaluation was normal.
2.1.3 Patient 3
He is a 3-years-old male child, second in birth order, born of third degree consanguineous couple (Figure 1). The child had severe global developmental delay, with onset of nystagmus at age 2 months and generalized seizures at age 7 months. His weight at the time of examination was 10 kg (−2 SD), height 90 cm (−1 SD), and head circumference, 42 cm (−5 SD). He had microcephaly, spasticity, dystonia, and brisk deep tendon reflexes. Brain imaging was suggestive of microcephaly, bilateral cerebral atrophy, leukoencephalopathy in frontotemporal regions predominantly, cystic changes and calcifications in frontotemporal white matter, enlarged centricles, severe brain stem atrophy (Figure 1(1a)). BERA (brainstem evoked response audiometry) revealed normal hearing. Pendular nystagmus was noted on ophthalmology evaluation suggestive of cortical visual blindness.
His elder male sibling (patient 4) had similar manifestations and expired at 3 years of age due to lower respiratory tract infection. The age at presentation of the symptoms was 4 months. He presented with global developmental delay and conculsions. He was born at term by normal vaginal delivery. He was noticed to be irritable and had poor suck reflex. He had no head holding at 7 months of age. He had nystagmus, microcephaly, dystonia, spasticity, brisk deep tendon reflexes. Brain imaging was suggestive of multiple punctate calcifications in the basal ganglia and cortical white matter, severe white matter abnormalities in frontal and anterior temporal white matter with cystic changes in the periventricular white matter and frontal lobes. (Figure 1(4b)). Nystagmus was noted on ophthalmology evaluation. Cerebrospinal fluid (CSF) and serum interferon alpha levels done at 1-year of age was found to be absent in CSF and 26.22 pg/ml in serum. Lymphocytosis (>300 cells) was observed on CSF examination of elder child.
Detailed clinical phenotypes of all the four affected children are given in Table 1. The study has the approval of institutional ethics committee. Written informed consents were obtained for study participants.
| Individual | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|
| Origin | Indian | Indian | Indian | Indian |
| Consanguinity | + | + | + | + |
| Gender | Male | Male | Male | Male |
| Age at assessment | 6 months | 18 days | 3 years | 6 months |
| Age of presentation | 3 months | At birth | 7 months | 5 months |
| Sequencing | Whole-exome sequencing | Whole-exome sequencing | Targeted Sanger sequencing | − |
| Coding DNA change | c.205C>T | c.205C>T | c.205C>T | NA |
| Amino acid change | p.R69W | p.R69W | p.R69W | NA |
| Zygosity | Homozygous | Homozygous | Homozygous | NA |
| Exon | 2 | 2 | 2 | NA |
| Birth weight (gm/SD) | 2,500 (−2) | 1,800 (−3) | 2,750 (−1) | 2,700 (−1) |
| Anthropometry | ||||
| Weight (gm/SD) | 4,600 (−2) | 1,900 (−3) | 10,000 (−2) | NA |
| Length (cm/SD) | 59 (−4) | 48 (0) | 90 (−1) | NA |
| OFC (cm/SD) | 37 (−7) | 32 (0) | 42 (−3) | NA |
| Microcephaly | + | − | + | − |
| Seizures | + | + | + | + |
| Neurodevelopmental delay | + | + | + | + |
| Feeding difficulty | − | − | + | + |
| Dystonia | − | + | + | + |
| Tone | Spasticity | Spasticity | Spasticity | Spasticity |
| Deep tendon reflexes | Brisk | Brisk | Brisk | Brisk |
| Brain imaging | ||||
| Delayed myelination | + | + | + | + |
| Cystic changes | + | − | + | + |
| White matter abnormalities | + | + | + | + |
| Thin corpus callosum | + | + | + | + |
| Intracranial calcifications | + | + | + | + |
| Other investigations | ||||
| USG abdomen | Hepatomegaly | Splenomegaly, altered echotexture of liver | NA | NA |
| EEG findings | NA | Left temporal sharp waves | Epileptiform activities | Nonspecific: generalized background attenuation with low amplitude of normal sleep architecture |
| Echocardiography | NA | Patent ductus arteriosus | NA | NA |
| Ophthalmological findings | Nystagmus | Normal | Nystagmus, cortical visual blindness | Nystagmus |
| Hearing evaluation | NA | Normal | Normal | NA |
| Hematology | Normal | Normal | NA | NA |
| Thyroid stimulating hormone | NA | Elevated (22 mU/L) | NA | NA |
| Serum alpha-interferon (24–98 pg/ml) | NA | NA | NA | 26.22 |
| CSF alpha-interferon (pg/ml) | NA | NA | NA | Absent |
| CSF lymphocytosis | NA | NA | NA | 6 cells/mm3 |
- +present, −absent, NA, not available; OFC, occipitofrontal circumference; SD, standard deviation; USG, Ultrasonography; EEG, electroencephalography; CSF, cerebrospinal fluid; TSH, thyroid stimulating hormone.
2.2 Molecular testing
After obtaining informed consent, genomic DNA was extracted from the leukocytes by standard phenol-chloroform method. Whole-exome sequencing was carried out for patient 1 and patient 2. Validation and segregation analysis was carried out by Sanger sequencing. Sanger sequencing of the recurrent mutation was done for patient 3 and his parents.
2.2.1 Whole-exome sequencing
Whole-exome sequencing (WES) was carried out as described previously to achieve an average coverage depth in the range of 100–130×, such that ∼95% of the bases are covered at >20×, with a sensitivity of >90% (Girisha et al., 2016). WES raw data was processed using SeqMule v1.2.5 and the called variants were annotated by ANNOVAR v.2016Feb01 (Guo, Ding, Shen, Lyon, & Wang, 2015; Wang, Li, & Hakonarson, 2010). The overall variant filtering strategy is outlined in Supplementary Table S2. Variants with minor allele frequency less than or equal to 1% and met the PHRED quality criteria (>20) were retained. The synonymous variants were excluded and the homozygous variants lying within the distance of 20 base-pairs from the exon were included for further analysis. The homozygous variants other than the one in RNASEH2C were not considered as they were not correlating with the observed phenotype.
2.2.2 Sanger sequencing
Primers were designed for exon 2 of RNASEH2C (NM_032193.3) gene and immediate flanking intronic sequences using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). Primers are available upon request. Using the genomic DNA, exon 2 was amplified by Polymerase Chain Reaction. Sanger sequencing was performed on the purified PCR amplicons using ABI PRISM 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
2.2.3 Homozygosity mapping
Using FILTUS software (Department of Medical Genetics, University of Oslo, Norway), homozygosity mapping was performed for patient 1 and patient 2 with default input file settings after loading the VCF files (Danecek et al., 2011). The parameters for minimum segment size and posterior threshold were set to 2 and 0.5 Mb, respectively for running the AutEx algorithm (Vigeland, Gjotterud, & Selmer, 2016).
3 RESULTS
All four individuals in our study were born to consanguineous families from western and southern parts of India. Clinical findings of all four patients are summarized in Table 1. Cystic changes in the brain were noted in patient 1, patient 3, and patient 4. Hepatomegaly and splenomegaly were noted in patient 1 and patient 2, respectively. Patient 2 was observed to have elevated levels of thyroid stimulating hormone. The homozygous variant, c.205C>T (NM_032193.3, p.Arg69Trp) was identified in all the three families in this study. Molecular analysis could not be carried out for patient 4 as the DNA samples were not available due to his early demise.
Regions of homozygosity was identified using FILTUS software in patient 1 and 2 who underwent exome sequencing (Supplementary Table S3) (Crow et al., 2006). A common 2.3 Mb region of homozygosity (ROH) in chromosome 11 containing RNASEH2C was found (Supplementary Table S1). Patient 1 and patient 2 shared the same haplotype, “A-T-A-A” in this 2.3 Mb region. These single nucleotide polymorphisms were Sanger sequenced in patient 3 who showed the presence of three out of the four SNPs, T-A-A in the haplotype (chr11:64940713C>T, chr11:65487856G>A, chr11:67266279G>A).
4 DISCUSSION
Early onset encephalopathy, seizures, neurodevelopmental delay, spasticity, delayed myelination, hypoplastic corpus callosum, white matter abnormalities, intracranial calcifications were the presenting features of our patients as observed in the reported literature (Crow et al., 2015). Chilblain lesions, usually found in 40% of the cases with AGS was not observed in our patients (Rice et al., 2007). Hypothyroidism has been observed in 14% of the patients with TREX1 mutations. However, this is the first-time hypothyroidism is noted in patients with AGS type 3. No significant intrafamilial variability was observed in patient 3 and patient 4 as reported earlier in one of the families with this founder mutation (Vogt et al., 2013).
In a study reporting 374 patients from 299 families, 12% of the AGS patients have been found to be due to biallelic mutations in RNASEH2C gene. A total of 37 families with mutations in RNASEH2C have been reported till date. Currently, nine mutations are identified in RNASEH2C including seven missense, one splice site variation, one small indel and one small deletion (http://www.hgmd.cf.ac.uk/ac/all.php, public version accessed on 10 April 2017). Twenty-five of these families are of South Asian origin and homozygous for p.Arg69Trp mutation in RNASEH2C. Currently, all the patients with this mutation are of Indian and Pakistani origin. This variant is found to contribute to 72% of the cases with AGS type 3. Other variations are observed in single families (Crow et al., 2015; Ramantani et al., 2010).
The variant, c.205C>T has an allele frequency of 0.0006822 in South Asian population as observed in gnomAD browser and is reported in three patients of Indian origin in literature (Merchant, Verma, Shah, Kulkarni, & Jalan, 2016; Ramantani et al., 2010; Ramesh, Sankar, Gowrishankar, & Ganapathy, 2013). The carrier frequency of the allele is not known in our population. Another truncating variant, c.58dup, in addition to this founder mutation is also reported in an Indian family (Ramesh et al., 2013). By haplotype mapping in five Pakistani families, Crow et al. identified the founder effect for this variant. The same variant was observed in all the three affected probands in our study. Hence, further analysis was carried out to investigate the founder effect. The common haplotype, “A-T-A-A,” spanning 2.3 Mb region on chromosome 11 signifies the possibility of a founder effect in them. This haplotype was retrospectively validated in patient 3, who shared three of these single nucleotide polymorphisms, T-A-A. This implicates the notion that c.205C>T mutation represents a founder mutation.
In conclusion, our study adds to the clinical spectrum of AGS type 3 and reports three families from India with the founder mutation, supporting the previous studies on RNASEH2C. Considering our findings and review of literature, we suggest to screen South Asian children with Aicardi–Goutieres syndrome for this variant as the first step in molecular diagnostic testing strategy.
ACKNOWLEDGMENT
We thank the families who cooperated with the evaluation of the subjects and consented for participation in this study. This work was supported by Department of Health Research funded the project titled “Clinical and molecular characterization of leukodystrophies in Indian children” (V.25011/379/2015-GIA/HR).
CONFLICTS OF INTEREST
None.