Homozygous single base deletion in TUSC3 causes intellectual disability with developmental delay in an Omani family
Abstract
Intellectual disability (ID) is the term used to describe a diverse group of neurological conditions with congenital or juvenile onset, characterized by an IQ score of less than 70 and difficulties associated with limitations in cognitive function and adaptive behavior. The condition can be inherited or caused by environmental factors. The genetic forms are heterogeneous, with mutations in over 500 known genes shown to cause the disorder. We report a consanguineous Omani family in which multiple individuals have ID and developmental delay together with some variably present features including short stature, microcephaly, moderate facial dysmorphism, and congenital malformations of the toes or hands. Homozygosity mapping combined with whole exome next generation sequencing identified a novel homozygous single base pair deletion in TUSC3, c.222delA, p.R74 fs. The mutation segregates with the disease phenotype in a recessive manner and is absent in 60,706 unrelated individuals from various disease-specific and population genetic studies. TUSC3 mutations have been previously identified as causing either syndromic or non-syndromic ID in patients from France, Italy, Iran and Pakistan. This paper supports the previous clinical descriptions of the condition caused by TUSC3 mutations and describes the seventh family with mutations in this gene, thus contributing to the genetic spectrum of mutations. This is the first report of a family from the Arabian peninsula with this form of ID. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Intellectual disability (ID), previously called mental retardation, is an early-onset neurodevelopmental disorder that impacts on daily living skills. An affected individual struggles with memory, problem solving, language and visual comprehension as well as daily living skills such as self-care, independence and interpersonal communication. These deficits are reflected by an IQ (intelligence quotient) score of less than 70 [Salvador-Carulla et al., 2011; American Psychiatric Association, 2014]. ID affects 1–3% of the general population [Maulik et al., 2011] and can be caused by genetic or environmental factors. Environmental causes include problems with pregnancy or childbirth, post-natal toxicity, malnutrition, infection or injury. There is considerable genetic heterogeneity underlying familial ID (syndromic or not) [Najmabadi et al., 2011]. Chromosomal abnormalities can account for up to 15% of these cases [Miller et al., 2010]. There are also over 500 genes that have been linked to ID [Sun et al., 2015] including genes with de novo mutations in isolated cases [Hamdan et al., 2014]. The proteins that are disrupted in ID consist of components of the neuronal synaptic complex as well as molecules involved in vesicle trafficking and exocytosis, cell–cell and cell–matrix interactions, glutamate receptors and their downstream effectors, the Rho and ERK/MAPK pathways, zinc finger transcription factors and chromatin remodelling [Kaufman et al., 2010].
In this report, we describe clinical features and molecular analysis in a consanguineous Omani family with multiple affected members that have ID with developmental delay caused by a novel TUSC3 homozygous mutation. Biallelic TUSC3 mutations have previously been shown to cause both non-syndromic and syndromic ID [Garshasbi et al., 2008; Molinari et al., 2008; Garshasbi et al., 2011; Khan et al., 2011; Loddo et al., 2013; El Chehadeh et al., 2015]. These findings support a presumed role of normal TUSC3 in the glycosylation of neurodevelopmental proteins [Kelleher and Gilmore, 2006; Mohorko et al., 2014].
MATERIALS AND METHODS
The family was contacted and ascertained through the Genetic and Developmental Medicine Clinic at the Sultan Qaboos University Hospital, Muscat, Oman. This study was approved by the Sultan Qaboos University Ethical Committee. The affected individuals were recruited to the study with the informed consent of their parents using a process that adhered to the tenets of the Declaration of Helsinki. Peripheral blood was sampled from each individual by venipunture for genomic DNA extraction by the Qiagen spin column method (Qiagen Ltd., Manchester, UK) according to standard procedures. Ethnically matched Omani controls were recruited from students of the Leeds Omani Society, using a process approved by the Leeds Central Research Ethics Committee (reference number 08/H1313/17). Control DNAs were sampled using Oragene saliva collection tubes (DNA Genotek Inc., Ontario, Canada) and genomic DNA was extracted using the manufacturer's protocol. Array comparative genomic hybridization (array-CGH) was performed using the Agilent Human Genome CGH 105A arrays according to the supplier's instructions (Agilent Technologies Limited, Wokingham, UK). Whole-genome homozygosity mapping was performed using Affymetrix SNP 6.0 arrays (AROS Biotechnologies Ltd., Aarhus, Denmark) and the data analyzed and presented using AgileMultiIdeogram software (http://dna.leeds.ac.uk/autozygosity/). Whole exome next generation sequencing was carried out using the SureSelect Target Enrichment V.4 reagent (Agilent Technologies Limited) for library preparation, followed by deep sequencing using paired end reads on an Illumina HiSeq 2500 (Illumina Incorporation, Little Chesterford, UK) according to the manufacturer's protocols. The sequencing reads were aligned to the human genome reference hg19 using Bowtie2 software and processed in the SAM/BAM format using Picard and the Genome Analysis Toolkit. Annovar software (http://www.openbioinformatics.org/annovar/) was used to annotate the variants. To check for homozygous regions in the exome data, the sequence variant list was analyzed using AgileVariantMapper and presented as a multi-Ideogram as described above (http://dna.leeds.ac.uk/autozygosity/). In looking for pathogenic variants of disease significance, any variants with a read depth of less than 10 were excluded, as were variants lying outside the exons and flanking splice site regions, synonymous variants, variants with a minor allele frequency > 1% in the exome variant server or 1,000 genomes databases as well as heterozygous variants. For PCR and Sanger sequencing, oligonucleotide primers were designed with Primer3 (http://primer3.ut.ee/). PCR was performed on genomic DNA through thermal cycling with an initial denaturing step of 95°C for 2 min then 35 cycles of 94°C for 30 sec (denaturing), 65°C for 30 sec (annealing) and 72°C for 45 sec (extension) followed by a final extension step of 72°C for 5 min. The products were treated with ExoSAP-IT (GE Healthcare, Freiburg, Germany) and sequenced using the BigDye terminator v.1.1 cycle sequencing kit (Applied Biosystems, Darmstadt, Germany) on an ABI3130xl Genetic Analyzer (Applied Biosystems). Results were analyzed using Sequence Analysis v.5.2 software (Applied Biosystems).
RESULTS
Clinical Features
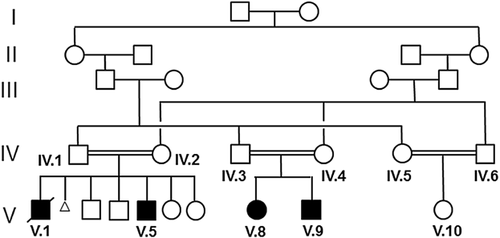
There were four patients with ID as the consensus feature that were identified in the pedigree (Fig. 1 and Table I). Patient V.5, a male, was initially referred to the clinic at 7-years old for short stature (below the 3rd centile) and developmental delay of language and motor skills. In school, V.5 has difficulties with reading and writing. The patient also had a myopathic elongated face with a pointed chin, downslanting palpebral fissures, apparent hypertelorism, a downturned mouth with thin lips as well as very thin nails and bilateral 2–3 syndactyly of the toes. In view of the child's features which are reminiscent of Smith–Lemli–Opitz syndrome, 7-dehydrocholesterol was tested and found to be within normal limits. Further investigations including chromatography of plasma amino acids, urine organic acids, lactate, ammonia and creatinine kinase were all within normal reference ranges. Magnetic resonance imaging (MRI) of the brain was normal. His behavior included a fascination with the sight and sound of water. Family history revealed that his deceased older brother (V.I) also had similar clinical features as well as camptodactyly of the right fingers which were confirmed by reading his hospital notes and reviewing family pictures. He had also been assessed by the developmental team and was found to fulfil the DSM-5 diagnostic criteria for autism spectrum disorder (ASD). Patient V.I drowned in water at the age of 10. Patients V.8 and V.9, a female and male, respectively, share a similar developmental and clinical profile as patient V.5 and his deceased brother (see Table I), though patient V.9 does not have any congenital malformations of the hands or toes. Patient V.8 has shown some delay of motor functions including problems with balance and a poor hand grip and was unable to hold a pencil between the thumb and finger, resulting in her being unable to draw a circle or a straight line. She has had difficulty chewing food and swallowing, and sometimes drools when performing a focused task. Her communication comprises a few single words, vowel sounds and non-verbal gestures including pointing and facial expressions. It is worth highlighting that the unaffected female sibling V.10 also appears to have some of the facial features described in the affected patients, so the mild dysmorphic features described in this pedigree may not be disease-specific.
| ID (Gender) | V.1 (M) | V.5 (M) | V.8 (F) | V.9 (M) | V.10 (F) |
|---|---|---|---|---|---|
| TUSC3 genotype (status) | M/M (affected) | M/M (affected) | M/M (affected) | M/M (affected) | M/+ (unaffected) |
| Growth parameters (at birth) | Wt = 50th centile | Wt = 3rd centile | Wt = 3rd centile | Wt = 3rd centile | Wt = 90th centile |
| Ht = 10th centile | Ht = 10th centile | Ht = < 3rd centile | Ht = < 3rd centile | Ht = 50th centile | |
| HC = 25th centile | HC = 3rd centile | HC = < 3rd centile | HC = < 3rd centile | HC = 50th centile | |
| Congenital malformations | Unilateral camptodactyly—hand | Bilateral 2–3 syndactyly—toes | Bilateral 2–3 syndactyly—toes | − | − |
| Growth parameters (age) | Wt = 3rd centile | Wt = < 3rd centile | Wt = 50th centile | Wt = 3rd centile | Wt = 3rd centile |
| Ht = < 3rd centile | Ht = < 3rd centile | Ht = 10th centile | Ht = < 3rd centile | Ht = 10th centile | |
| HC = 3rd centile | HC = 3rd centile | HC = 25th centile | HC = < 3rd centile | HC = 50th centile | |
| (9 years) | (7 years) | (3 years) | (1 year) | (5 years) | |
| Facial features | Hypertelorism | Hypertelorism | Hypertelorism | Hypertelorism | Hypertelorism |
| Long face | Long face | Thin philtrum | Thin philtrum | ||
| Deep set eyes | Thin philtrum | ||||
| Thin philtrum | Pointed chin | ||||
| Pointed chin | |||||
| Brain imaging | N/R | Normal MRI | N/R | N/R | Normal MRI |
| IQ | N/R | 53 | 40 | 50 | 81 |
| Autism spectrum disorder | + | − | − | − | + |
| Developmental profile | N/R | Motor and language skills delayed | Motor and language skills delayed | Walked at 20 months | Walked at 19 months |
| Poor reading and writing ability | Can say ∼5 words with meaning at 1 year | Single words at 2.5 years | |||
| Current language normal but has delayed echolalia; also has hyperlaxia |
- Growth parameters were measured using the World Health Organisation child growth standards and IQ was measured using the Stanford Binet intelligence test (5th edition). Wt, weight; Ht, height; HC, head circumference; N/R, not recorded; +, presence; and −, absence.
Molecular Analysis
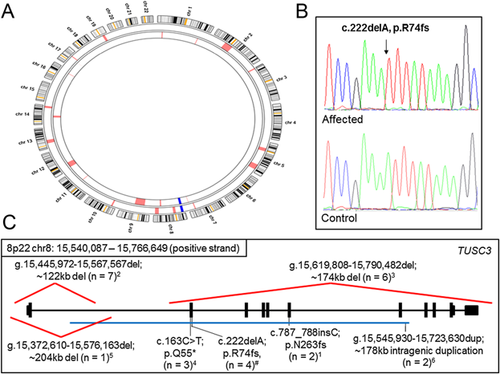
In order to determine whether a chromosomal rearrangement was the cause of ID, comparative genome hybridization (array-CGH) was performed in V.5, who showed no chromosomal rearrangement. To locate the pathogenic mutation in this family, genomic DNA from V.8 and V.9 was initially analyzed. These individuals have similar manifestations to their affected cousins V.1 and V.5, and so are likely to harbor the same causative genetic mutation. Analysis of DNA from V.8 by homozygosity mapping highlighted several regions, any of which could harbor the causative mutation (Fig. 2A). These included regions on chromosomes 2, 3, 5, 7, 8, 10, 13, 14 and 19. To identify the mutation, whole exome next generation sequencing was performed on V.9. An initial list of 61,905 variants was used to check for homozygous regions. Combining the single nucleotide polymorphism data of V.8 with the whole exome sequencing data from V.9 highlighted a single ∼11.5 Mb homozygous region on chromosome 8 common to both affected individuals. To look for the pathogenic mutation, the initial variant list was reduced to 113 following variant filtration, and this was further reduced by excluding those variants that lay outside the common homozygous region on chromosome 8. This left only one candidate, a novel variant in TUSC3 (tumor suppressor candidate 3) [MIM 610385] which is a known ID gene. Sanger sequencing confirmed the presence of this single base deletion, which causes a frameshift mutation (NM_006765/NM_178234:c.222delA:p.R74 fs) (Fig. 2B). This mutation segregated in all the affected cases in the family as expected for recessive inheritance. The mutation was absent from 60,706 subjects on the ExAc database (http://exac.broadinstitute.org/, February 2015), and was also not present in 50 ethnically matched Omani control DNAs, providing further support as to the scarcity of this allele in the human population.
DISCUSSION
TUSC3, also known as N33 or OST3A (oligosaccharyl transferase 3 homolog A), encodes a 348 amino acid protein containing a signal peptide, membrane anchored thioredoxin fold and four transmembrane domains. The protein is localized to the endoplasmic reticulum, where it forms part of the multimeric oligosaccharyl transferase complex that is likely to be involved in post-translational glycosylation of selected proteins which are essential for normal brain development [Kelleher and Gilmore, 2006; Mohorko et al., 2014]. It has been suggested that TUSC3 may be a magnesium transporter [Zhou and Clapham, 2009], though the results of that study could also be explained by the ability of TUSC3 to modulate the glycosylation efficiency of such a membrane transporter. It has also previously been observed that the chromosome 8p22 genomic region containing TUSC3 is frequently deleted in somatic cells in prostate [Bergerheim et al., 1991] and ovarian cancers [Pribill et al., 2001]. Furthermore, silencing TUSC3 in ovarian cancer cell lines enhanced proliferation and migration, suggesting that the protein may be a tumor suppressor [Vaňhara et al., 2013]. The affected patients described in this study have not developed tumors, but all are relatively young and it may be appropriate to monitor for increased tumor risk in the future.
Here, we describe a novel null mutation in TUSC3 causing ID with dysmorphic features in a consanguineous family from Oman. This is the seventh family that has been identified with TUSC3 mutations causing syndromic or non-syndromic ID (Fig. 2C). Previous reports have identified a premature stop codon mutation [Garshasbi et al., 2011], frameshift insertion [Molinari et al., 2008], gross deletion [Garshasbi et al., 2008; Khan et al., 2011; Loddo et al., 2013], and an intragenic duplication [El Chehadeh et al., 2015], all of which would be expected to produce no functional TUSC3 protein product. The mutation reported herein is a frameshift mutation in exon 2, which may lead to nonsense mediated decay of the RNA transcript, or may give rise to a truncated, non-functional protein. It is interesting to note that primary fibroblast cell cultures derived from patients with a null homozygous mutation and controls showed no significant differences in glycosylation of selected proteins suggesting that TUSC3 may not have a major role in the glycosylation of extracerebral proteins [El Chehadeh et al., 2015]. Given the spectrum of TUSC3 mutations observed in ID cases and families, it seems likely that the condition is caused by lack of functional TUSC3 protein during neurodevelopment rather than build-up or activity of defective mutated protein.
Previous reports [Garshasbi et al., 2008; Molinari et al., 2008; Garshasbi et al., 2011; Khan et al., 2011; Loddo et al., 2013; El Chehadeh et al., 2015] have shown that patients with biallelic TUSC3 mutations have moderate to severe ID with speech delay as the prominent features. These findings are consistent with the patients described in this report. Additional characteristics observed in some of the reported cases included short stature, microcephaly and moderate facial dysmorphism, and these signs were also present in some of our patients. It is worth highlighting that three of the four patients studied here also had congenital malformations of the toes or hands, which have been seen before in a single patient who also had unilateral cryptorchidism [Loddo et al., 2013]. Overall, there appears to be no obvious genotype-phenotype correlation between type of mutation and the presence of additional features [El Chehadeh et al., 2015].
This is the first time that mutations in this gene have been described in the Arab population. The family could be expected to benefit from this knowledge through genetic counselling and also through carrier screening of additional family members and/or potential marriage partners. Omani genetic services, which are government funded, have been developed and improved significantly within the healthcare system over the last 10 years [Rajab et al., 2013]. This coincided with a growing understanding of the increased burden of recessive conditions and congenital abnormalities due to consanguinity, with over 50% of marriages in this population being consanguineous [Rajab and Patton, 2000; Rajab et al., 2013]. Although consanguinity is rooted in the culture and reflects the preservation of traditions, religion and tribal groups, genetic health education and the culturally sensitive implementation of technologies such as prenatal screening and pre-implantation genetic diagnosis may offer some hope for the family described in this report.
To summarize, we have identified a novel TUSC3 mutation in a consanguineous family from Oman. Our findings contribute further data on the spectrum of mutations causing this form of ID and the phenotype associated with them.
ACKNOWLEDGMENTS
We would like to thank the families who participated in this study for their full cooperation. The study was funded by a scholarship awarded to AAA by the Oman Ministry of Higher Education.