Early detection of bilateral cataracts in utero may represent a manifestation of severe congenital disease
Abstract
We observed bilateral cataracts on second trimester ultrasound, in two consecutive pregnancies, with no other structural defects detected. The parents were unrelated and had no family history for the disease. The first pregnancy was terminated in week 22. Copy number variation analysis revealed, in both the aborted fetus and the mother, a 495 kb duplication at 22q11.23 encompassing CRYBB3 and CRYBB2, and not present in variation databases. In the second pregnancy, lens hyperechogenicity was detected by ultrasound at week 13 and 4 days. The identical duplication at 22q11.23 was found in the fetus and considered as possibly pathogenic. At weeks 22 and 30, smaller orbit measurements were elucidated on ultrasound, raising concerns as to the underlying molecular genetic cause, necessitating further investigation. Whole-exome sequencing, using DNA of the first fetus, was performed shortly after the birth of a male child, and two truncating RAB3GAP1 mutations were detected: c.538G>T; p. (Glu180*) and c.943C>T; p. (Arg315*). Neither mutation has been previously reported to be disease-causing; however, evaluation in the context of previously published literature indicated their deleterious nature, implying a clinical diagnosis of Warburg micro syndrome or Martsolf syndrome. Sanger sequencing confirmed segregation of the two mutations within the family, consistent with autosomal recessive inheritance. The child born from the second pregnancy showed features typical of Warburg micro syndrome, with the exception of microcephaly, at age 31 months. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Cataracts inherited as a monogenic trait are a clinically and genetically heterogeneous group of disorders. Disease-causing mutations in almost 200 genes have been identified, including syndromic forms, which have been estimated to represent nearly 20% of hereditary bilateral congenital cataracts [Rahi and Dezateux, 2000; Shiels et al., 2010].
Warburg micro syndrome (WARBM) is an autosomal recessive disease associated with mutations in four genes; RAB3GAP1 (WARBM1; OMIM #600118), RAB3GAP2 (WARBM2; OMIM #614225), RAB18 (WARBM3; OMIM #614222), and TBC1D20 (WARBM4; OMIM #615663) [Aligianis et al., 2005; Bem et al., 2011; Borck et al., 2011; Handley et al., 2013; Liegel et al., 2013]. It is a very severe disorder characterized by developmental delay, typically with no milestones attained beyond 4 months of age. Other features of WARBM include postnatal onset microcephaly, polymicrogyria, hypogenesis of the corpus callosum, cerebellar and cerebellar vermis hypoplasia, severe truncal hypotonia, and progressive limb spasticity leading to quadriplegia. In male patients, hypogonadism is common. Poor visual acuity has been attributed mainly to progressive optic atrophy and cortical impairment. In addition, bilateral congenital cataracts, microphthalmia, microcornea, and atonic pupils are typically present [Warburg et al., 1993; Aligianis et al., 2005; Bem et al., 2011; Borck et al., 2011; Handley et al., 2013; Liegel et al., 2013].
In one family, RAB3GAP1 mutations were described in association with Martsolf syndrome (OMIM #212720) [Handley et al., 2013]; a less severe condition characterized by congenital cataracts and microphthalmia, but without optic atrophy or cortical visual impairment, less pronounced microcephaly and intellectual disability, and a progressive spasticity that may be confined to the lower limbs [Martsolf et al., 1978; Handley and Aligianis, ; Handley et al., 2013].
In this report, we describe the clinical and laboratory investigations undertaken to determine the molecular pathogenesis of bilateral cataracts, detected in utero by ultrasound, in two consecutive pregnancies.
CLINICAL REPORT
The research was conducted in accordance with the Declaration of Helsinki, with appropriate Institutional Review Board approval prior to the start of the study, and informed consent obtained from study participants.
A 27-year-old primigravida of white Czech origin was referred for routine pregnancy screening at a local tertiary care centre—Gennet (Prague). The father was of white Slovak origin. There was no reported consanguinity, nor family history of monogenic disorders.
Fetal nuchal translucency thickness, serum free beta-hCG, and PAPP-A in the first trimester did not reveal an increased risk for aneuploidy of chromosomes 13, 18, and 21. The second trimester screen involved a high-resolution ultrasound anomaly scan, using the Voluson E8 (General Electric Medical Systems, Kretztechnik GmbH & Co., Zipf, Austria), with transabdominal and transvaginal high-frequency probes. Hyperechogenic lenses were noted bilaterally at week 20 and 5 days. No other developmental abnormalities were observed. The head circumference was at the 82nd centile [Chitty et al., 1994]. The rest of the growth, the amount of amniotic fluid, and Doppler parameters were also normal. Amniocentesis was performed at week 20 and 6 days. The karyotype performed on standard G-banded metaphases from amniotic fluid cultivation was 46 XY, the TORCH screen was negative, as was testing for 7-dehydro-cholesterol in the amniotic fluid.
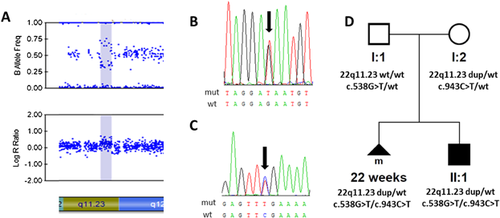
Copy number variation (CNV) analysis revealed a maternally inherited 495 kb microduplication at 22q11.23 encompassing six protein-coding genes; arr[hg19] 22q11.23(25,124,711–25,620,142)x3 (Fig. 1A), including CRYBB2 (OMIM *123620) and CRYBB3 (OMIM *123630)—2 genes known to be implicated in the pathogenesis of non-syndromic autosomal dominant cataract [Graw, 2009]. No prior entry for microduplication of this size existed in public CNV databases, nor in 1,600 population-matched controls. A report outlining the cytogenetic findings was issued; however, the family was lost to subsequent follow-up.
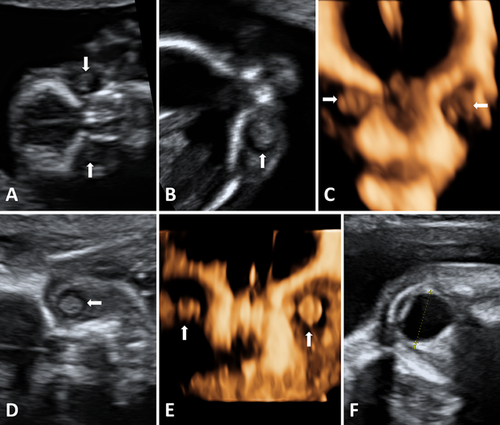
At age 29 years, the mother was referred with a second pregnancy. Reportedly, the first pregnancy was terminated in week 22, and no pathology, except for bilateral cataracts, was found at the autopsy. Combined first-trimester screening did not show any indication of chromosomal abnormalities. High-resolution ultrasound scan at 13 weeks and 4 days gestation revealed that bilateral lens hyperechogenities were already present (Fig. 2A–C), and confirmed as cataracts at 16 weeks and 4 days (Fig. 2D and E).
Amniocentesis in the second pregnancy had been performed at week 17. The same microduplication arr[hg19] 22q11.23(25,124,711–25,620,142)x3, present in both the mother and the terminated fetus, was detected (Fig. 1A and D), leading to the hypothesis that it may be responsible for the cataract phenotype, due to altered expression of CRYBB2 and CRYBB3.
The parents decided to continue with the pregnancy. Subsequent ultrasound scans at 19 weeks and 5 days and at 22 weeks and 5 days found a symmetrically growing fetus with no other organ anomalies, and with a head circumference corresponding to the 33rd and 71st centiles, respectively [Chitty et al., 1994]. The orbital diameters (measured in the transverse section of the fetal orbital region, from the mid-echogenicities of the lateral and medial orbital margins) at 22 weeks was 9.2 mm (<10th centile of fetal ocular nomograms), while the transverse diameter of the lenses (measured in the coronal plane) was 4.4 mm (50th centile). At 30 weeks, these measurements were 12.1 mm (<10th centile) and 4.8 mm (10th centile), respectively (Fig. 2F) [Goldstein et al., 1998]. Dilated ocular examination of the mother did not reveal any lens opacities.
The detection on serial scans, of orbital diameters below the 10th centile in the growing fetus during the second pregnancy, had raised further concerns regarding the underlying molecular pathology. Both parents were also very keen for molecular genetic investigation, to permit the opportunity for avoiding the pathology in subsequent pregnancy. As the crystalline lenses of the mother were clear, and microduplications spanning CRYBB2 and CRYBB3 had not been reported as causing cataracts, we decided to perform whole-exome sequencing (WES) using DNA derived from cultured cells from amniotic fluid of the aborted fetus.
The second pregnancy resulted in a male child, born spontaneously at 36 weeks and 5 days of gestation. The weight at birth was 2,700 g and height 47 cm. Head circumference was 33 cm, corresponding to the 50th centile for the gestational age [Barbier et al., 2013]. The presence of cataracts in both eyes was confirmed immediately after birth. In addition, bilateral cryptorchidism was noted. Developmental delay, truncal hypotonia, and increased muscle tone in the legs were noted at age 4 months. Head circumference at age 7 months was 42.75 cm (10th–25th centile). At age 12 months the infant weighted 8,440 g (10th centile), and his length was 69.5 cm (<5th centile) [Vignerová et al., 2006]. Other systemic signs and symptoms present at 1 year included truncal hypotonia, micropenis, developmental delay (i.e., inability to crawl), poor visual fixation, low-set, posteriorly angulated ears, beak nose, long philtrum, mild hypertelorism, and hyper-extended first toes, but no limb contractures. At 14 months of age, the head circumference reached 45.75 cm and at age 31 months, 48 cm (both values at the 10th centile) [Vignerová et al., 2006]. The infant did not have feeding problems. He could not crawl or turn at age 31 months and his speech was limited to monosyllables.
Cataract surgery was performed at age 2 months in the left eye, and 3 months in the right eye. A poor pupillary dilating reaction to cycloplegic drops was noted. At age 13 months, the corneal horizontal diameter was 9.75 mm in the right eye and 10.00 mm in the left eye, consistent with the diagnosis of microcornea [Muller and Doughty, 2002]. Temporal pallor of the optic head nerves was also found.
Using WES, several rare sequence variants were identified but only two in RAB3GAP1, c.538G>T; p. (Glu180*), and c.943C>T; p. (Arg315*) (reference sequence NM_012233.2), were predicted to be pathogenic based on the published literature [Handley et al., 2013] and frequency in public variant databases (Supplemental Table SI). Sanger sequencing validated the presence of both mutations in RAB3GAP1 in the fetus from the first pregnancy, and segregation in the parents (Fig. 1D). With the onset of developmental delay, both RAB3GAP1 mutations were subsequently confirmed in the child born from the second pregnancy (Fig. 1B and C).
MOLECULAR INVESTIGATION
Fetal DNA from both pregnancies was extracted from cultured cells in amniotic fluid, using the QIAamp DNA micro kit (Qiagen GmbH, Hilden, Germany) [Becvárová et al., 2011]. Parental DNA was extracted from venous blood lymphocytes, with the MagNA Pure Compact Nucleic Acid Isolation Kit (Roche, Basel, Switzerland).
CNV analysis was performed on both fetal and parental DNA samples using a genome-wide SNP array HumanCytoSNP-12v2.1 BeadChip Kit (Illumina, Inc., San Diego, CA) containing ∼300,000 markers. Labeling and hybridization were performed according to manufacturer's instructions, with scanning undertaken on the Illumina iScan. Results were analyzed by CNVPartition 3.2.0 software tool within GenomeStudio v2011.1 (Illumina). Identified CNVs were mapped against human genome assembly UCSC hg19 (http://www.genome.ucsc.edu).
Distinction of possibly pathogenic from benign CNVs occurring as normal genomic variation was based on the search in the DGV database (http://projects.tcag.ca/variation), the DECIPHER database v9.1 (http://decipher.sanger.ac.uk), and by comparison with internal data set comprising more than 1,600 Czech unrelated individuals (unpublished data).
WES was performed using SureSelect Human All Exon 50 Mb Kit (Agilent, Berkshire, UK) by the 100-bp paired-end method on the Illumina HiSeq2000 instrument according to the manufacturer's protocol (fee for service; Axeq Technologies, Rockville, MD). Coverage at the raw data level was at least 75-fold. 94.4% of target regions had a coverage depth of more than 10-fold. Raw FASTQ reads were aligned against UCSC hg19 version of the human genome, using the Burrows-Wheeler Alignment tool (http://bio-bwa.sourceforge.net/) [Li and Durbin, 2009]. Variant calling was performed with SAMTOOLS (http://samtools.sourceforge.net/) [Li et al., 2009].
Sequence variants within genes known to be associated with both non-syndromic and syndromic human cataracts (as listed on http://cat-map.wustl.edu/, accessed September 2015) [Shiels et al., 2010] were prioritized for further assessment. Frequency of the identified variants was determined from the Exome Aggregation Consortium (ExAC, Cambridge, MA, http://exac.broadinstitute.org), version 0.3 and the Exome Variant Server (EVS, http://evs.gs.washington.edu/EVS/), version: 0.0.25. Conventional Sanger sequencing was used to verify the presence of possibly pathogenic mutations, and to determine their segregation within the family. Primers and conditions are available upon request.
DISCUSSION
The benefits of next-generation sequencing for the diagnosis of genetically heterogeneous diseases are undisputable. To the best of our knowledge, using WES and achieving a molecular diagnosis in the case of isolated ocular pathology observed prenatally has not yet been described. Our study highlights the importance of careful observation of the eyes/lenses during prenatal anatomic survey, for which high-resolution sonographic visualization is necessary.
The preliminary results from WES, showing the likely presence of two truncating mutations in RAB3GAP1, were available shortly after the birth of a male son from the second pregnancy. After confirmation by Sanger sequencing and segregation analysis within the family, we have interpreted that the predicted deleterious nature of the two identified mutations, in the context of previously published literature, suggested a clinical diagnosis of either WARBM or Martsolf syndrome. The postnatal clinical course, including the child's morphological features, were consistent with the diagnosis of WARBM, apart from the absence of microcephaly at 31 months. It has been recently suggested that disease associated with RAB3GAP1, and other genes causing WARBM and Martsolf syndrome, may in fact represent a continuous phenotypic spectrum [Handley et al., 2013], which is in line with the clinical findings of the patient presented in the current study.
Including the current study, pathogenic RAB3GAP1 mutations in a compound heterozygote state have been reported in only eight families [Handley et al., 2013]. The recent improvements in CNV detection have also necessitated a better understanding of their role in phenotypic diversity. As the mother carrying the arr[hg19] 22q11.23(25,124,711–25,620,142)x3 microduplication was clinically unaffected, we do not believe this particular CNV has an influence on the phenotype in the affected child. It should be, however, noted that the functional impacts of interactions between point mutations and other structural variants are currently poorly understood [Henrichsen et al., 2009].
In clinical settings, a custom-designed target enrichment permitting parallel analysis of known disease-causing genes is often being used as an alternative to WES. With panels, the genes are usually more fully covered and application of this approach in patients with congenital cataracts has recently revealed a detection rate of 75% [Gillespie et al., 2014]. WES, however, in conditions with uncharacterized genes, such as hereditary cataracts, permits the data to be revisited and reassessed when novel genes are reported, in addition to the opportunity to more comprehensively exclude an additional Mendelian disorder.
Identification of the molecular genetic cause creates several possible implications for future pregnancies of the couple. Although cataracts could be seen on early ultrasound screening in both previous pregnancies, we consider either preimplantation genetic diagnosis, or invasive testing with targeted analysis, to be more reliable in avoiding recurrence of the disease. The final decision should be, however, made by the couple after informed discussion.
In summary, our study shows the clinical utility of WES, using a fetal sample, to determine the molecular pathogenesis of ocular anomalies identified on prenatal ultrasound. In the absence of other structural abnormalities, fetal lens hyperechogenicities may still indicate a serious condition like WARBM. WES can allow early diagnosis, reducing the period of diagnostic uncertainty associated with expensive, time-consuming clinical testing. Predicting the phenotypic outcome of novel variants remains, however, a challenge.
ACKNOWLEDGMENTS
The research was supported by a grant from Nadace Leontinka, UNCE 204011, and PRVOUK-P24/LF1/3 programs of the Charles University in Prague. LD was supported by SVV 260148/2015.