Novel splicing mutation in the ASXL3 gene causing Bainbridge–Ropers syndrome
Abstract
Bainbridge–Ropers syndrome (BRPS) is characterized by severe developmental delay, feeding problems, short stature, characteristic facal appearance including arched eyebrows and anteverted nares, and ulnar deviation of the hands. BRPS is caused by a heterozygous mutation in the additional sex combs-like 3 (ASXL3) gene. We describe a patient with severe developmental delay, feeding problems, short stature, autism, and sleep disturbance with a heterozygous de novo splicing mutation in the ASXL3 gene. Reported disease-causing mutations in ASXL3 are located mostly in the first half of exon 11, analogous to ASXL1 mutations of which result in Bohring–Opitz syndrome (BOS). Our findings suggest that the expression of the truncated ASXL3 protein, including ASXN and ASXH domains, give rise to BRPS, which is distinct from but overlaps with BOS. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Bainbridge–Ropers syndrome (BRPS: OMIM #615485) is a recently identified syndrome of severe developmental delay, feeding problems, short stature, ulnar deviation of hands, and characteristic facal changes of arched eyebrows and anteverted nares. BRPS is caused by a heterozygous mutation in the additional sex combs-like 3 (ASXL3) gene on chromosome site18q12 [Bainbridge et al., 2013]. Six cases of this syndrome have been reported so far [Bainbridge et al., 2013; Dinwiddie et al., 2013; Zhu et al., 2015]. All reported pathogenic mutations in ASXL3 have been either frameshift or truncating mutations. De novo truncating mutations in ASXL1, which belongs to the same gene family as ASXL3, cause Bohring–Opitz syndrome (BOS), which shares clinical manifestations with BRPS.
We describe a patient with severe developmental delay, feeding problems, short stature, and autism, with a heterozygous de novo splicing mutation in the ASXL3 gene.
This study was approved by the institutional review board of Nagoya City University Graduate School of Medical Sciences.
PATIENT
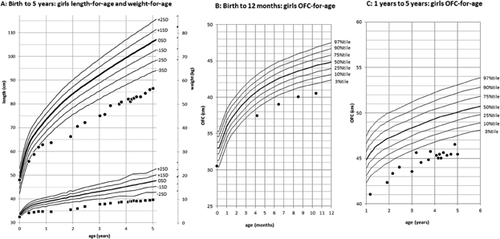
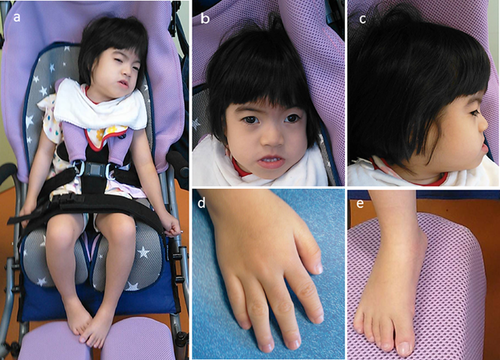
The patient is a 5-year-old Japanese girl with unremarkable family history. She is an only child. The girl was born normally at the 38th gestational week without any perinatal abnormalities. At birth she had a weight of 2,430 g (−0.9 SD), a length of 48.0 cm (−0.1 SD) and an occipital frontal circumference (OFC) of 30.5 cm (−1.8 SD). After birth she was hospitalized for about two weeks because of feeding difficulties and inadequate weight gain. Feeding difficulties gradually improved, but developmental delay became apparent. She was able to hold her head up by 4 months, sat without support at 2 years, and stood holding on to things at 3 years. At 4 years she only spoke a few words. She has shown an abnormal sleep pattern since her infancy. She would stay up all night and sleep during the day. The use of ramelteon (melatonin receptor agonist) was only partially effective. After birth she showed failure to thrive (Fig. 1). At 5 years she had a weight of 9.6 kg (−3.4 SD), a length of 86.5 cm (−4.7 SD) and an OFC of 45.4 cm (−3.0 SD). She had a prominent forehead, thick eyebrows, long lashes, exotropia, depressed nasal ridge, thin upper lip vermillion, hirsutism, microcephaly, bilateral camptodactyly of third, fourth and fifth fingers, deep palmar creases, and small hands and feet (Fig. 2). Deep tendon reflexes and muscle tone were normal. She had astigmatism and hyperopia. Results of head MRI studies, plasma amino acids, urine organic acids, blood gas analysis, lactate, and pyruvic acid were normal. Conventional G-banded chromosome analysis showed a 46,XX karyotype. At 5 years her childhood autism rating scale (CARS) score for autism was 35.5, indicating mild to moderate autism.
MATERIALS AND METHODS
We performed whole-exome sequencing on the patient and her parents. The genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Midi Kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). Three micrograms of DNA was sheared into 150–200-bp fragments using the Covaris LE220 or M220 Focused-ultrasonicator (Covaris, Woburn, MA). To capture the exonic DNA, we used the SureSelect XT Human All Exon V5 capture library (Agilent Technologies, Santa Clara, CA). We then constructed a sequence library using the SureSelect XT Target Enrichment System for Illumina Paired-End Sequencing Library kit (Agilent Technologies), and performed DNA sequencing of 100-bp paired-end reads using the Illumina HiSeq 2000 sequencer. The sequencing data were mapped to a reference genome (GRCh37/hg19) using BWA (ver.0.6.1), and the resulting average read depth of the targeted regions was 67.5. Variant calling was performed using SAMtools (ver.0.1.16) and GATK (ver.1.6) software. To identify disease-causative mutations, we excluded all known variants found in public databases (dbSNP142, 1,000 Genomes Project, Exome Aggregation Consortium (ExAC) and NHLBI ESP6500) and a control in-house database (221 controls of normal Japanese individuals and with other diseases), except for those also identified as pathogenic mutations in the NCBI ClinVar and HGMD databases. We focused on non-synonymous SNVs, insertions and deletions (indels), and splice-site variants.
RESULTS
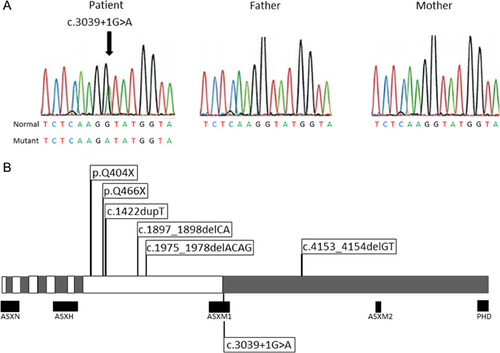
In the ASXL3 gene (NM_030632), we identified a heterozygous single nucleotide mutation (c.3039 + 1G>A) in the invariant “GT” splice donor site 3′ of exon 11, which was absent in her parents, indicating that the mutation arose de novo. The finding was confirmed by Sanger sequencing (Fig. 3A). The mutation is predicted to have dramatic consequences on the maturation process of the ASXL3 mRNA leading to the removal of exon 11, which would create a frame shift and truncate the protein. In addition, compound heterozygous mutations were discovered in CPAMD8 (NM_015692; c.5185C>T; p.R1729C and c.218C>A; p.A73E). These were not confirmed by Sanger sequencing because CPAMD8 was deemed irrelevant to the phenotype of the patient.
DISCUSSION
Here we report the first case of BRPS caused by an ASXL3 splicing mutation. The splice donor site mutation is predicted to affect the sequence region that is important in the recognition of the intron/exon boundary and removal of the intron, resulting in the skipping of the adjunct exon or activation of a cryptic splice site [Berget, 1995]. BRPS-causing mutations reported to date, as well as the one reported here, are listed in Figure 3B. These include frameshift and nonsense mutations, and now a splicing mutation. These mutations would result in the expression of a truncated protein, retaining exons one through 10 including ASXN and ASXH domains [Katoh, 2013], and could be an underlying pathomechanism of BRPS.
Recently, de novo truncating mutations in ASXL1 have been shown to account for approximately 50% of cases with BOS [Hoischen et al., 2011]. ASXL1 and ASXL3 belong to the same gene family. BRPS shares clinical features with BOS such as poor feeding and developmental delay. It is of note that observed BOS-causing mutations in ASXL1 are also de novo heterozygous truncating mutations due to nonsense or frameshift mutations. Additionally, known disease-causing nonsense mutations in ASXL1 occur almost exclusively within a very limited region of the protein. Disease-causing mutations in ASXL3 are truncating mutations, and are located mostly in the first half of the exon 11, which is analogous to that in ASXL1, where mutations resulting in BOS occur [Hoischen et al., 2011]. No pathogenic missense mutations have been reported in ASXL1 or ASXL3. And all truncating mutations in ASXL1 and ASXL3 would retain ASXN and ASXH domains. Therefore, our findings suggest that the truncating ASXL3 protein including ASXN and ASXH domains have toxic effects as in the truncating ASXL1 protein, and give rise to BRPS, which is distinct from but overlaps with BOS.
Our patient showed clinical features of BRPS including severe developmental delay, feeding problems, postnatal growth retardation as in Table I. BRPS and BOS share several clinical features, whereas some are distinct (Table I) [Hastings et al., 2011; Bainbridge et al., 2013; Dinwiddie et al., 2013]. To date, none of the patients with ASXL3 mutations have displayed the typical BOS posture (defined as having three out of four features: external rotation and/or adduction of the shoulders, flexion at the elbows, flexion at the wrists, and ulnar deviation of the wrists and/or fingers at the metacarpophalangeal joints [Hastings et al., 2011]). Our patient did not have BOS posture either. Because all of the 30 reported patients with ASXL1 mutations show the typical BOS posture, this feature could be useful to distinguish the two “related” conditions [Magini et al., 2012]. Five of the eight patients with BOS reportedly showed sleep disturbances [Russell et al., 2015]. The patient with BRPS described by Dinwiddie et al. [2013], and our patient also showed impaired sleep organization. Sleep disorder may be common in BOS and BRPS.
| Our patient | BRPS (Bainbridge et al., 2013 and Dinwiddie et al., 2013) | BOS (Hastings et al. 2011) | |
|---|---|---|---|
| Clinical | |||
| Feeding difficulties | + | 4/5 | 14/14 |
| IUGR | + | 2/5 | 11/14 |
| Severe/profound developmental delay | + | 5/5 | 14/14 |
| Recurrent infections | − | NA | 11/14 |
| Seizures | − | NA | 6/14 |
| Craniofacial | |||
| Microcephaly | + | 4/5 | 14/14 |
| Trigonocephaly | − | 1/5 | 12/14 |
| Micro/retrognathia | + | 1/5 | 13/14 |
| Nevus flammeus | − | 0/5 | 12/14 |
| Prominent eyes | − | NA | 14/14 |
| Abnormal palate | − | 3/5 | 10/14 |
| Hypertelorism | − | 2/5 | 9/14 |
| Upslanting palpebral Fissures | − | NA | 7/14 |
| Depressed nasal bridge | + | NA | 7/14 |
| Anteverted nares | − | 4/5 | 8/14 |
| Low-set posteriorly angulated ears | − | 3/5 | 8/14 |
| Ophthalmic | |||
| Strabismus | + | 1/5 | 8/14 |
| Myopia | − | 1/5 | 8/14 |
| Hair/skin | |||
| Low hairline | − | NA | 7/14 |
| Hypertrichosis | + | 2/5 | 5/14 |
| Neurological/skeletal | |||
| BOS posture | − | 0/5 | 14/14 |
| Fixed contractures | + | NA | 8/14 |
- +, present; −, absent; IUGR, intrauterine growth restriction (birthweight <10th centile); NA, not applicable.
- Bold-suggested diagnostic criteria of BOS.
ACKNOWLEDGMENTS
This study was supported in part by a grant for Research on Applying Health Technology from the Ministry of Health, Labour and Welfare of Japan to F. M., N.O., M.K., M.Y., K.Y., K.K., and S.S. We thank Mr. K.A. Boroevich for English proofreading.