Dual genetic diagnoses: Atypical hand-foot-genital syndrome and developmental delay due to de novo mutations in HOXA13 and NRXN1
Abstract
We describe a male patient with dual genetic diagnoses of atypical hand-foot-genital syndrome (HFGS) and developmental delay. The proband had features of HFGS that included bilateral vesicoureteric junction obstruction with ectopic ureters, brachydactyly of various fingers and toes, hypoplastic thenar eminences, and absent nails on both 4th toes and right 5th toe. The atypical features of HFGS present were bilateral hallux valgus malformations and bilateral preaxial polydactyly of the hands. Chromosomal microarray analysis identified a de novo 0.5 Mb deletion at 2p16.3, including the first four exons of the NRXN1 gene. Whole exome sequencing and subsequent Sanger sequencing identified a de novo missense mutation (c.1123G>T, p.Val375Phe) in exon 2 of the HOXA13 gene, predicted to be damaging and located in the homeobox domain. The intragenic NRXN1 deletion is thought to explain his developmental delay via a separate genetic mechanism. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Hand-foot-genital syndrome (HFGS [MIM 140000]) is a rare autosomal dominant limb and urogenital malformation syndrome. It is characterized by bilateral thumb and great toe hypoplasia (due to a short distal phalanx and/or 1st metacarpal or metatarsal). Hallux varus is a useful diagnostic sign when present. Urogenital abnormalities in males include hypospadias, ectopic ureteric orifices, vesicoureteric reflux (VUR) and pelviureteric junction obstruction [Stern et al., 1970; Verp et al., 1983; Cleveland and Holmes, 1990]. Developmental milestones and intellect are usually normal. Post-axial polydactyly was reported in a large autosomal dominant pedigree (MIM 176305) [Guttmacher, 1993]. However, preaxial polydactyly does not appear to have been previously reported in HFGS.
HFGS is caused by mutations (missense, nonsense, and polyalanine expansions) in the homeobox transcription factor gene, HOXA13 [Mortlock and Innis, 1997; Goodman et al., 2000], although there are a few reported patients with clinical features of HFGS without an identified mutation in HOXA13 [Goodman et al., 2000]. Chromosome 7p15.2 deletions encompassing HOXA13 have also been reported as causing HFGS [Devriendt et al., 1999; Hosoki et al., 2012]; these patients also displayed intellectual impairment likely due to haploinsufficiency of other genes in addition to HOXA13.
Neurexins are a family of polymorphic neuronal cell adhesion molecules and receptors that mediate synapse formation and signalling. The neurexin 1 (NRXN1) gene has two independent promoters generating two major isoforms, neurexin 1-α and neurexin 1-β. Deletions involving NRXN1 are associated with a wide spectrum of neuropsychiatric disorders, including developmental delay (particularly speech), autism, schizophrenia and mild dysmorphic features; but they have not been associated with limb or urogenital anomalies [Ching et al., 2010; Schaaf et al., 2012; Béna et al., 2013; Dabell et al., 2013].
We present an atypical case of HFGS in a male patient with preaxial polydactyly, hallux valgus and development delay where the clinical presentation is likely due to a novel HOXA13 missense mutation and an intragenic NRXN1 deletion.
CLINICAL REPORT
The 4-year-old male proband is the only child born to healthy non-consanguineous white Australian parents. The parents had no developmental or medical issues and there was no significant family history. He was seen by clinical genetics at 2 years and 8 months of age due to developmental delay along with limb and renal anomalies. In the pregnancy, a 20-week fetal anomaly scan identified bilateral hydronephrosis, but no other anomalies. He was born at 35 weeks gestation via a normal delivery with a birth weight of 3.11 kg (>90th centile) [Kitchen et al., 1983]. Postnatal imaging demonstrated hydronephrosis due to bilateral vesicoureteric junction (VUJ) obstruction and ectopic ureters. He required staged ureterostomies for VUJ obstructions and recurrent febrile urinary tract infections. He was also noted to have digital anomalies and had bilateral inguinal hernia repairs. There were no concerns regarding his feeding, growth, behavior, hearing, or vision. His development was delayed, as he crawled at 15 months, walked at 21 months, and at the age of 2 years and 8 months had just started running, had a palmar grasp of objects, and had only 20 single words. He was otherwise in good health.
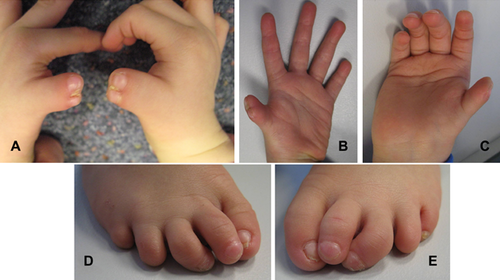
On examination at 2 years and 8 months, his growth parameters were all within normal limits (weight 13 kg [25th centile], height 91 cm [25th centile], and head circumference 51.4 cm [90th centile]). He had bilateral preaxial polydactyly of the hands (bifid thumbs); brachydactyly of both thumbs, 2nd and 5th fingers; clinodactyly of both 5th fingers; hypoplastic thenar eminences; bilateral hallux valgus; brachydactyly of both 2nd, 4th, and 5th toes; bilateral 3rd toe clinodactyly; and absent nails on both 4th toes and the right 5th toe (Fig. 1).
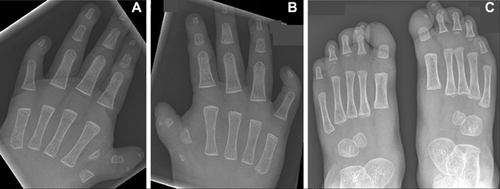
A skeletal survey revealed bilateral thumb duplication, short proximal and distal phalanges of the medial left thumb, short proximal and absent distal phalanges of right thumb, blunting of the distal phalanges of both index fingers, absent middle phalanges of both 5th fingers, and terminal resorption of the distal phalanges of all toes (Fig. 2).
METHODS
Written informed consent for this study was obtained from both parents. The proband and both parents had DNA extracted from peripheral blood (NucleoBond CB100, Macherey-Nagel, Germany) for the purposes of SNP microarray and whole-exome sequencing studies.
SNP Microarray Studies
SNP microarray was performed using the Illumina HumanCytoSNP-12 version 2.1 [Bruno et al., 2011]. All procedures for fragmentation, labeling and hybridization were performed at the Australian Genome Research Facility (Melbourne, Australia) and were done according to the manufacturer's protocol (Illumina, San Diego, CA). Raw data was processed using Karyostudio (Illumina, San Diego, CA) and probe intensity measurements were normalized to a reference set of 100 local clinical samples (samples referred as part of routine genetic investigations for developmental delay, autism and behavioral disorders). The effective resolution was approximately 200 kilobases.
The clinical significance of Copy Number Variants (CNVs) greater than 200 kilobases (Victorian Clinical Genetics Services reporting threshold based on 20 probes required for each call, with an average probe density of one probe per 10 kilobases) was determined by comparison with public databases of copy number variants (i.e., Children's Hospital of Philadelphia [CHOP], DatabasE of Genomic variants and Phenotype in Humans Using Ensembl Resources [DECIPHER], Database of Genomic Variants [DGV], and International Standards for Cytogenomic Arrays [ISCA]), as well as with an internal database of CNVs observed in 55,000 clinical samples. The UCSC genome browser March 2006 NCBI GRCh36/hg18 assembly was used for comparison of genomic coordinates detected for each copy number variant called at the laboratory reporting threshold.
Whole Exome Sequencing Studies
Whole exome sequencing (WES) was performed as previously reported [Tsurusaki et al., 2014]. Briefly, 3 µg of genomic DNA was sheared using the Covaris S2 system (Covaris, Woburn, MA) and captured using a SureSelect Human All Exon V5 (50 Mb) library (Agilent Technologies, Santa Clara, CA). Captured DNA was sequenced using HiSeq 2000 (Illumina, San Diego, CA) with 101 bp paired-end reads and 7 bp index reads. Image analysis and base calling were performed by sequence control software real-time analysis and CASAVA software v1.8.2 (Illumina). Mapping to the human reference genome (UCSC hg19, NCBI builds 37.1) was performed using Novoalign 3.00.02. After the removal of PCR duplication by Picard, the single nucleotide variants (SNVs) and short insertions and deletions (Indels) were called using Genome Analysis Toolkit (GATK) 1.6–5. Called SNVs and Indels were annotated using ANNOVAR (2013 Jun).
Out of all variants within exons or ±2 bp from the exon–intron boundaries, we removed synonymous variants, those within the segmental duplications, and those registered in all dbSNP137, the Human Genetic Variation Database (HGVD), the National Heart Lung and Blood Institute Exome Sequencing Project Exome Variant Server (NHLBI-ESP 6500), and our in-house databases (exome data from 575 Japanese individuals).
The variants were subsequently validated by Sanger sequencing using an ABI3500xl or ABI3100 autosequencer (Life Technologies, Carlsbad, CA) with the following primers (forward: 5′-AAGAAGCGCGTGCCTTATAC-3′; reverse: 5′-CTCCTTCGGGAGAGGAAAAT-3′). The polymerase chain reaction (PCR) was performed in 10 µl containing 10 ng of genomic DNA, 10 × Ex Taq Buffer, dNTP mixture (2.5 mM each), primer mixture (final conc. 1.0 µM each), and Ex Taq (5 units/µl) (Takara Bio, Inc., Otsu, Japan) using a T1 Thermocycler (Biometra, Göttingen, Germany) with the following program: preincubation (98°C for 2 min); amplification by touchdown PCR (denaturation at 94°C for 30 sec, annealing at 66–60°C (step-down, 2°C/cycle) for 30 sec and extension at 72°C for 45 sec, for 40 cycles).
RESULTS
SNP Microarray Studies
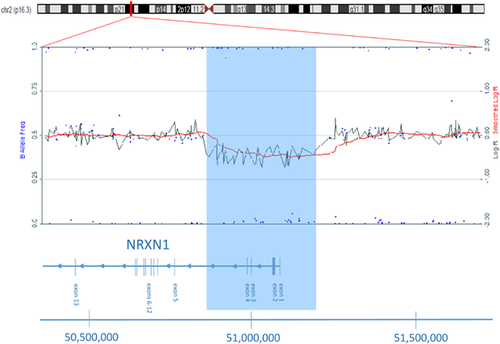
SNP microarray analysis in the proband identified an approximately 0.5 Mb deletion at chromosome region 2p16.3. The deleted region, which contained 46 SNP probes, involved genomic co-ordinates chr2: 50,797,032–51,283,384 (NCBI GRCh36/hg18), and included exons 1–4 of the α-isoform of the NRXN1 gene (Fig. 3). Microarray analyses on parental samples were normal, indicating this to be a de novo finding.
Comparison of the Copy Number Imbalance Within NRXN1 Using Bioinformatics Tools
Comparison of the deleted region in this patient to other copy number imbalances involving the NRXN1 gene was carried out using publicly available web-based bioinformatics tools (CHOP, DECIPHER, DGV, ISCA, OMIM, and Pubmed). There is some recorded benign copy number variation in the DGV, but there are numerous other case reports in the literature of whole gene or intragenic deletions of NRXN1.
Whole Exome Sequencing Studies
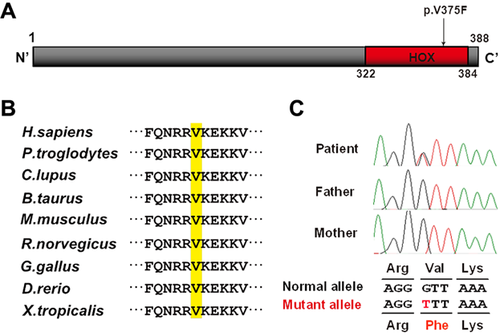
We performed trio-based WES. The mean depth of protein-coding regions of RefSeq genes was 83.37–100.80 reads, with 91.4–93.2% of target sequences covered by 20 or more reads. Only one novel mutation (c.1123G>T, p.V375F) in exon 2 of HOXA13 remained as a candidate variant after excluding non-pathogenic variants as described in the methods, based on the autosomal dominant model. The mutation had prediction scores of damaging or disease causing by SIFT, PolyPhen2 and MutationTaster, and occurred in the homeobox domain at a highly evolutionally conserved amino acid among several species from zebrafish to humans (Fig. 4a and b). Sanger sequencing of DNA from the patient and his parents confirmed that the mutation has occurred de novo (Fig. 4c).
DISCUSSION
The proband in this report presented with developmental delay, severe urogenital anomalies and limb anomalies, including preaxial polydactyly. Subsequent genetic investigations revealed two separate mutations in NRXN1 and HOXA13. The intragenic NRXN1 deletion is thought to have contributed to the neurodevelopmental difficulties seen in this patient and the HOXA13 missense mutation is thought to have caused the limb and urogenital anomalies. Patients with dual genetic diagnoses are difficult to recognize clinically, although their identification has increased due to the widespread use of microarray technology and massively parallel sequencing. Yang et al. [2013] found that 4 out of 250 patients referred for whole-exome sequencing received two non-overlapping molecular diagnoses. Similarly, Farwell et al. [2015] found that 11 out of 500 patients received dual molecular diagnoses.
The neurexins are a family of cell-adhesion molecules and receptors that are located on the neuronal cell surface and mediate synapse formation and signalling. Human neurexins are encoded by three unlinked genes (NRXN1, NRXN2 and NRXN3). Haploinsufficiency of NRXN1 due to whole gene or exonic deletion has been associated with a spectrum of neuropsychiatric and developmental disorders, including autistic spectrum disorder (ASD), intellectual impairment, hypotonia and motor developmental delay, language delay, epilepsy, schizophrenia, and minor dysmorphic features [Ching et al., 2010; Schaaf et al., 2012; Béna et al., 2013; Dabell et al., 2013]. Neurexin deletions show incomplete penetrance and may be found in healthy individuals [Ching et al., 2010; Schaaf et al., 2012; Béna et al., 2013; Dabell et al., 2013]. Significant urogenital or limb anomalies do not appear to have been reported before in association with NRXN1 deletions and are most likely accounted for by the presence of the HOXA13 mutation in this patient.
Homeobox genes encode transcription factors that regulate cell proliferation and differentiation during morphogenesis. Hox genes clusters control the polarity of the embryo. There are four Hox gene clusters (Hoxa, Hoxb, Hoxc, Hoxd) and their physical position on the chromosome seems to correlate with the time and place of expression [Goodman, 2002]. HOXA13 is the most distal member of the HOXA gene cluster and is involved with distal limb and genitourinary development [Du and Taylor, 2004]. HFGS is currently the only disorder associated with mutations in HOXA13.
The pattern of limb malformations in HFGS is characterized by bilateral thumb and great toe hypoplasia with hallux varus. The limb anomalies appear to be fully penetrant and symmetrical. The genitourinary phenotype is variable and incompletely penetrant. To date, 4 missense, 4 nonsense and 10 polyalanine expansion mutations have been reported [Mortlock and Innis, 1997; Goodman et al., 2000; Innis et al., 2002; Frisen et al., 2003; Innis et al., 2004; Utsch et al., 2007; Jorgensen et al., 2010; Parker et al., 2011; Owens et al., 2013; Imagawa et al., 2014]. A more severe phenotype (including absent halluces, postaxial polydactyly and nail anomalies) has been observed associated with missense mutations in the homeodomain [Goodman et al., 2000; Innis et al., 2002; Parker et al., 2011; Imagawa et al., 2014], and it is postulated that this is via a different mutational effect [Imagawa et al., 2014]. Table I summarizes the mutation spectrum and clinical features of reported patients with severe HFGS.
| “Classic” HFGS Stern et al. [1970]; Mortlock and Innis [1997] | Goodman et al. [2000] (family 5) | Guttmacher [1993]; Innis et al. [2002] | Parker et al. [2011] | Imagawa et al. [2014] | Current case | |
|---|---|---|---|---|---|---|
| Sex | M | Father, son, daughter | F | F | M | |
| HOXA13 variant | N372H | Q371L | R326G | I368F | V375F | |
| Located in homeobox domain | + | + | + | + | + | |
| Thumbs | Hypoplastic | Severely hypoplastic | Hypoplastic | Hypoplastic | Severely hypoplastic | Severely hypoplastic |
| Proximally placed | Proximally placed | Proximally placed | Bilateral duplication | |||
| Halluces | Hypoplastic/absent | Absent | Severely hypoplastic (variable) | Valgus deformity | Absent | Valgus deformity |
| Varus deformity | ||||||
| Nails | Normal | NR | Absent 1st/2nd toenails | Normal | Small/absent | Absent 4th/5th toenails |
| Thenar eminence | Hypoplastic | NR | NR | Hypoplastic | NR | Hypoplastic |
| Other limb features | Clinobrachydactyly of 5th fingers | Clinobrachydactyly of 5th fingers | Post-axial polydactyly | Thumb-index finger syndactyly | Severe, absent DIP creases | Clinobrachydactyly of 5th fingers |
| Ulnar deviation of 2nd fingers | Hypoplasia of all middle phalanges | Uniphalangeal 2nd toes | Brachydactyly of 2nd fingers | |||
| Hypoplasia of middle phalanges | Hypoplasia of all middle phalanges, and absent middle phalanges of 5th fingers | |||||
| Clinodactyly of 3rd toes | ||||||
| Male genitalia | Hypospadias | Hypospadias | Hypospadias | NA | NA | Normal |
| Female genitalia | Longitudinal vaginal septum, double uterus, double cervix | NA | NR | Double uterus Double cervix | Bicornuate and hypoplastic uterus | NA |
| Urinary abnormality | Ectopic ureteric orifices, vesicoureteric reflux, pelviureteric junction obstruction | NR | NR | Normal | Grade 3–4 hydrouretonephrosis | Hydronephrosis due to bilateral vesicoureteric junction obstruction and ectopic ureters |
| Growth | Normal | NR | Normal | Normal | Normal | Normal |
| Psychomotor development | Normal | NR | Normal | Normal | Normal | Delayed |
- KEY: +, present; NA, not applicable; NR, not recorded.
The presence of preaxial polydactyly in this patient is unusual and has not been reported before in patients with HFGS. Hallux valgus is also unusual, but has been reported before in one patient with an R326G missense mutation in the homeobox domain [Parker et al., 2011]. Postaxial polydactyly was reported in one family where affected individuals were later found to have a Q371L missense mutation in the homeobox domain [Guttmacher, 1993; Innis et al., 2002]. Clinically affected individuals in this family were also found to have a 2 bp deletion (−78–79delGC) in the gene's promoter region on the same allele. Other clinically unaffected family members were found to have only the 2 bp promoter deletion. It was therefore postulated that the 2 bp promoter deletion produced no detectable abnormalities on its own, but along with the Q371L missense mutation likely contributed to the unusual phenotype. A previous report concluded that the residue V375 is critical for structurally recognizing TAA in the major groove, and that HOXA13 dimerization is required to activate transcription of target genes [Zhang et al., 2011]. Thus, we hypothesise that the V375F mutation in our patient may cause a more severe and atypical phenotype due to altered DNA binding and the subsequent dysregulation of genes involved in limb and genitourinary patterning during embryogenesis.
CONCLUSION
This report highlights atypical upper and lower limb malformations of bilateral preaxial polydactyly and hallux valgus deformities in a patient with HFGS. The presence of polydactyly (pre- or post-axial), nail anomalies and/or hallux valgus in cases of HFGS may be due to specific missense mutations in the homeodomain of HOXA13, which confer a more severe phenotype. This patient also had neurodevelopmental difficulties likely explained by the presence of a NRXN1 intragenic deletion. This report also educates clinicians to search for other genetic diagnoses that may account for phenotypes not associated with the primary diagnosis (dual genetic diagnoses).
ACKNOWLEDGMENTS
We are particularly grateful to the parents who supported our publication of their child's case for the benefit of other families.
WEB RESOURCES
Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER), http://decipher.sanger.ac.uk
Database of Genomic Variants (DGV), http://projects.tcag.ca/variation
Ensembl, http://www.ensembl.org/index.html
HUGO Gene Nomenclature Committee (HGNC), http://www.genenames.org/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim
Pubmed, http://www.ncbi.nlm.nih.gov/pubmed/
NCBI genome remapping tool, http://www.ncbi.nlm.nih.gov/genome/tools/remap/




