Absence of skeletal anomalies in siblings with a maternally inherited 12q13.13-q13.2 microdeletion partially involving the HOXC gene cluster
Abstract
Microdeletions (12q13.13-q13.2) involving the HOXC gene cluster are rare. Only three patients with this contiguous deletion have been reported, all resulting in phenotypic features that include skeletal anomalies, facial dysmorphism, and intellectual disability. The deletion of the HOXC gene cluster is thought to result in skeletal anomalies in these patients. We report on siblings with a 969 kb deletion in the 12q13.13-q13.2 region detected by array comparative genomic hybridization (aCGH). This deletion spans seven of nine HOXC cluster genes. FISH analysis confirmed the siblings and mother were carriers of the 12q13.13-q13.2 deletion. Although minor facial dysmorphic features were present in both siblings, no skeletal anomalies were present in the siblings or the mother. The proband had autistic-like features and mild developmental delay, while the sibling and mother are of normal intelligence. The absence of skeletal anomalies in our family suggests that deletion of the entire HOXC gene cluster may be required to result in an abnormal skeletal phenotype, or those skeletal anomalies in previously reported patients may be attributed to other genes within the deletion interval. © 2014 Wiley Periodicals, Inc.
INTRODUCTION
HOX genes encode for a highly conserved group of transcription factors that play a significant role in patterning of the body axis during embryonic development [Suemori and Noguchi, 2000; Kmita and Duboule, 2003; Noordermeer et al., 2011]. In humans, a total of 39 HOX genes exist that are found within 4 clusters (A-D) located on chromosomes 7p15 (HOXA), 17q21.2 (HOXB), 12q13.13 (HOXC), and 2q31 (HOXD) [Lappin et al., 2006]. The critical properties of HOX gene clusters during embryogenesis include functional, spatial, and temporal collinearity [Durston et al., 2011]. Functional collinearity, the arrangement of genes in the same order as they act, is responsible for the HOX gene clusters role in body axis patterning. The spatial order and time sequence in which the HOX gene clusters are expressed, termed spatial and temporal collinearity, are unique phenomena that play a role in their genomic regulation. HOX gene cluster expression begins at the most 3′ gene, acting in the anterior portion of the embryo, and proceeds upstream to the most 5′ gene, acting in the posterior portion of the embryo [Durston et al., 2011].
Mutations or deletions involving individual HOX genes have been well documented to result in developmental disorders in both humans and mice, particularly skeletal anomalies [Goodman, 2002]. However, only three patients have been described involving hemizygous deletion of the HOXC gene cluster located in the 12q13.13 chromosome region. All three patients had microdeletions spanning the entire HOXC gene cluster and multiple other genes in the flanking region. These patients shared clinical features that include skeletal anomalies, facial dysmorphism, and intellectual disabilities [Okamoto et al., 2011; Jonsson et al., 2012; Hancarova et al., 2013].
Here, we report on a maternally inherited 12q13.13-q13.2 microdeletion spanning 7 of 9 HOXC genes in two siblings. Both siblings have mild facial dysmorphism, but do not display skeletal anomalies. The proband is a 4-year-old male with autistic-like features, mild developmental delay, facial dysmorphism, duplicated left ureter, and bilateral inguinal hernias. The sibling of the proband is a 7-year-old female with facial dysmorphism, short stature, and failure to thrive. Although the mother and daughter are deletion carriers, they both have normal intelligence.
CLINICAL REPORT
The proband is the second child of healthy and nonconsanguineous parents. He was born full term by an uncomplicated vaginal delivery to a 29-year-old, G2P1A0 (gravida 2, para 1, abortion 0) mother and a 30-year-old father. Prenatal ultrasound identified left hydroureteronephrosis. Birth weight was 4,200 g (75–90th centile) and length was 53 cm (75–90th centile). Renal and bladder ultrasound on Day 2 of life revealed severe left upper pole hydroureteronephrosis which was later found to be a double collecting system with the left ureter being dilated from the ureteropelvic junction to the ureterovesical junction. He underwent left upper pole heminephroureterectomy at 4 months of age. Bilateral inguinal hernias were notable after his second birthday and were repaired at 2 years 9 months.
He had features of autism spectrum disorder and mild developmental delay, sitting on time at 3–4 months of age, walking at 16 months, and first words at 14 months. Physical evaluation at 4 years of age identified arched eyebrows, deep set eyes, left epicanthal fold, over-folded ears, wide nasal bridge, cupid bow shape of the upper lip with smooth philthrum, high narrow palate, thin fingers with prominent fingertip pads and single-palmar creases bilaterally. He had normal muscle tone and no skeletal anomalies were identified. His weight was 16 kg (10–25th centile), height was 107 cm (25–50th centile) and head circumference was 50.2 cm (25–50th centile).
Family history is significant for a female sibling with history of failure to thrive and short stature. At 7 years of age her weight was 16 kg (<3rd centile), height was 42 cm (<3rd centile) and occipital frontal circumference (OFC) was 49.5 cm (10–25th centile). She had a mild motor delay, walking at the age of 16 months. Physical evaluation noted telecanthus, upslanting palpebral fissures, small nose, smooth philthrum, microretrognathia, abnormally shaped ears with a thick and overfolded helix and very prominent and enlarged antihelix, cutis marmorata, single prominent palmar creases bilaterally with very abbreviated superior crease, tapered fingers, long slender toes, with nails and toenails being minimally hypoplastic. She had normal tone and no evidence of skeletal anomalies. Renal ultrasound was not available. Other remarkable family history is poor weight gain as children for the mother and maternal grandmother, and several family members on the paternal side have mitochondrial encephalopathy with ragged red fibers (MERRF).
MATERIALS AND METHODS
Array comparative genomic hybridization (aCGH) analysis was performed on purified DNA from the siblings of our family using the SignatureChipO™ microarray, version 2.0 on a RocheNimbleGen 135K oligonucleotide array platform (Roche NimbleGen, Madison, WI), and scanned with a DNA Microarray Scanner (Agilent Technologies, Santa Clara, CA). Results were displayed by Genoglyphix v3.0 (Signature Genomic Laboratories, Spokane, WA). Fluorescence in situ hybridization (FISH) was performed on peripheral blood from the siblings and parents using a locus specific BAC probe. Images were captured using Isis FISH Imaging System v5.3 software (MetaSystems Group, Inc., Newton, MA). All aCGH and FISH procedures were performed according to manufacturer's protocol. The nucleotide coordinates are based on UCSC hg18 (NCBI Build 36, Mar. 2006).
RESULTS
Microarray analysis performed on the siblings DNA revealed a 969 kb heterozygous deletion (minimum size of deletion: 969 kb; maximum size of deletion: 1.01 Mb) in the 12q13.13-q13.2 region (chr12:52,653,352-53,622,392). This deleted interval contained 22 OMIM genes (HOXC11, HOXC10, MIR196A2, HOXC9, HOXC8, HOXC5, HOXC4, HOXC6, SMUG1, CBX5, HNRNPA1, NFE2, MIR148B, GPR84, ZNF385A, ITGA5, NCKAP1L, PDE1B, PPP1R1A, LACRT, DCD, MUCL1) and 10 non-OMIM genes (MIR615, FLJ12825, LOC100240735, LOC100240734, LOC400043, MIR3198-2, HNRNPA1P10, COPZ1, GTSF1, GLYCAM1). The nearest distal oligonucleotide probe (53,651,092-53,651,152) that is not deleted is approximately 28.7 kb away from the deletion region. The nearest proximal oligonucleotide probe (52,645,668–52,645,728) that is not deleted is approximately 7.62 kb away from the deletion region. The array karyotype for the siblings is arr[hg18] 12q13.13q13.2(52,653,352–53,622,392)x1. FISH analysis was performed on peripheral blood from the siblings for deletion confirmation using a BAC probe (RP11-753H16) located within the deleted region. In addition, FISH analysis was performed on peripheral blood from the parents to determine carrier status. The deletion was confirmed by FISH analysis in the siblings and mother. The FISH karyotype for the siblings and mother is nuc ish 12q13.1(RP11-753H16x1). The father was not found to carry the deletion by FISH analysis.
DISCUSSION
The two siblings in our study with a maternally inherited 12q13.13-q13.2 microdeletion spanning seven of nine HOXC genes are unique when compared to the three published patient reports involving de novo deletions of the entire HOXC gene cluster (Fig. 1). Clinical features of our 4-year-old male proband were autistic-like features, mild developmental delay, mild facial dysmorphism, duplicated left ureter, and bilateral inguinal hernias. The 7-year-old female sibling presented clinically with mild facial dysmorphism, short stature, failure to thrive and mild motor delay. Of importance, the mother and sibling of the proband were of normal intelligence, and family members with the deletion had no evidence of skeletal anomalies. Our proband had mild facial dysmorphic features that include a wide nasal bridge, smooth philtrum, and left-sided epicanthal folds. The sibling of the proband also had facial dysmorphic features that include mild telecanthus, microretrognathia, smooth philtrum, and upslanting palpebral fissures. The mother did not have facial dysmorphic features.
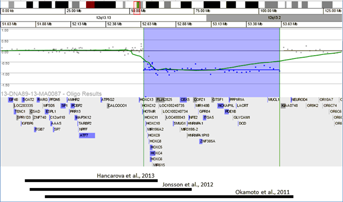
General facial anomalies have been reported in all previous published 12q13.13 deletion cases (Table I). Mutations or deletions of the HOXC genes have rarely been reported in humans. Only one patient with a homozygous nonsense mutation in the HOXC13 gene has been reported to present with pure hair and nail ectodermal dysplasia [Lin et al., 2012], while heterozygous carriers of the nonsense mutation were normal. Mouse studies have demonstrated that the HOXC gene cluster expression begins at the most 3′ gene, acting in the anterior portion of the embryo, and proceeds upstream to the most 5′ gene, acting in the posterior portion of the embryo [Durston et al., 2011]. The 3′ genes of the HOXC cluster are deleted in both siblings and the mother of our family (excluding HOXC12 and HOXC13). Facial dysmorphic features described in reported cases, including the siblings in our family, may be attributed to the deletion of 3′ HOXC cluster genes. However the mother in our family was not reported to have facial dysmorphic features. Deletion of these genes may have incomplete penetrance and variable expressivity.
| Features | Patient 1 [Okamoto et al., 2011] | Patient 2 [Jonsson et al., 2012] | Patient 3 [Hancarova et al., 2013] | Our family |
|---|---|---|---|---|
| Del size/chr region | 1.7 Mb (aCGH); 12q13.13-q13.2 | 1.13 Mb (aCGH); 12q13.13 | 0.9 Mb (SNP array); 12q13.13 | 969 kb (aCGH); 12q13.13-q13.2 |
| Location (hg18) | chr12:51,965,307-53,642,659 | chr12:51,834,791-52,971,391 | chr12:51,801,299-52,737,892 | chr12:52,653,352-53,622,392 |
| HOXC cluster involvement | 9 of 9 HOXC genes | 9 of 9 HOXC genes | 9 of 9 HOXC genes | 7 of 9 HOXC genes (excluding HOXC12 and HOXC13) |
| Inheritance | De novo | De novo | De novo | Maternal |
| Dysmorphic facial features | Long face, broad nose, prominent ears, bilateral low-set ears, downslanting palpebral fissures, strabismus, high palate | Bilateral epicanthic folds, depressed nasal bridge, slightly bulbous and antiverted nose, short philtrum | Long and narrow face, hypotelorism, enophthalmos wide nose | Proband: Wide nasal bridge, smooth philtrum, left-sided epicanthal folds |
| Sibling: Mild telecanthus, microretrognathia, smooth philtrum, upslanting palpebral fissures | ||||
| Mother: None | ||||
| Skeletal anomalies | Severe kyphosis, mild scoliosis, PIP joint flexion, contractures of 3rd and 4th fingers, inflexible DIP joints of index fingers, adducted thumbs on both hands | Short metacarpal and proximal phalangeal bones, congenital flexion contractures involving digits, hands, and elbows, ulnar deviation of both hands, valgus position of both ankles | Right hand flexion contractures of 4th and 5th fingers, conical shaped distal phalanges, ulnar deviation of both hands, disproportionate habitus (extremely long thorax, short lower limbs), hyperlaxicity of joints | Proband: None |
| Sibling: None | ||||
| Mother: None | ||||
| Intellectual disability | Expressive language and global development delay | Motor and mental development delay, attention deficit disorder | Mild intellectual disability | Proband: Autism spectrum Disorder, mild developmental delay |
| Sibling: None | ||||
| Mother: None | ||||
| Other | Tetrology of Fallot, bilateral inguinal hernias, hypodontia | Umbilical hernia | Ventricular septal defect, foramen ovale apertum, pulmonic stenosis, cryptorchidism, onychodystrophy, hyperelastic skin, abnormal palmar and plantar creases | Proband: Bilateral inguinal hernias, thin fingers with prominent fingertip pads |
| Sibling: Failure to thrive, tapered fingers, long slender toes |
- Chr: chromosome; Del: deletion; PIP: proximal interphalangeal; DIP: distal interphalangeal.
Common clinical features from published 12q13.13 deletion cases that are absent in our family include skeletal anomalies and severe intellectual disability (Table I). Okamoto et al. [2011 reported skeletal anomalies in a patient that included kyphoscoliosis, digital abnormalities (PIP joint flexion contractures of 3rd and 4th fingers, inflexible DIP joints of index fingers, adducted thumbs on both hands), and developmental delay. Jonsson et al. [2012 described a patient with skeletal anomalies that included digital abnormalities (short metacarpal and proximal phalangeal bones), congenital flexion contractures (involving digits, hands, and elbows), and intellectual disability. Lastly, the patient reported by Hancarova et al. [2013 had skeletal anomalies that included digital abnormalities (flexion contractures of 4th and 5th fingers on the right hand, conical shaped distal phalanges), abnormally long thorax, short lower limbs, and mild intellectual disability. None of the deletion carrying members in our family have skeletal abnormalities, however, the proband has thin fingers with prominent fingertip pads and his sibling has tapered fingers with long slender toes.
The most 5′ genes in the HOXC gene cluster region, HOXC12 and HOXC13, were not deleted in our patient, while all previously reported patients had deletions spanning the entire HOXC gene cluster (Fig. 1). Deletion of the HOXC cluster has been assumed to cause the skeletal anomaly [Okamoto et al., 2011]. If so, the absence of skeletal anomalies in our family could be explained by the incomplete deletion of the HOXC gene cluster. In addition, partial deletions or duplications of the HOXC cluster have been reported in DGV databases (DGV database updated July 30th, 2013, http://dgv.tcag.ca/dgv/app/home) as common structural variants (nsv899100: deletions involving HOXC4, HOXC5, HOXC6, HOXC8, HOXC9, HOXC10, HOXC11, and HOXC12; dgv512e1: deletions/duplications involving HOXC4, HOXC5, HOXC6, HOXC8, HOXC9, HOXC10, HOXC11, and HOXC12; nsv 899101 and nsv899102: deletions involving HOXC 4, HOXC4, HOXC5, HOXC6, HOXC8, HOXC9). Furthermore the possibility of later onset skeletal changes including kyphoscoliosis for our patient may also exist. Therefore, identification of this deletion will lead to continued clinical observations for skeletal changes in our patient. However, it is possible that the hemizygous deletion of the HOXC gene cluster alone may not cause skeletal anomalies seen in the published patients. Studies of the HOXC and HOXB clusters in mice suggest that a single set of HOX genes in one cluster is sufficient for embryonic body plan formation, and that genes of paralogous groups play complementary role(s) to each other [Suemori and Noguchi, 2000].
Moreover, the 12q13.13 deletion found in these patients varies greatly in size and involve multiple other genes in addition to the HOXC cluster. The SP7 gene is deleted in all three previously reported patients, while the RARG gene is also deleted in two of the three patients (Jonsson et al., 2012; Hancarova et al., 2013). The deletion detected in our family does not involve SP7 gene and RARG gene. The Sp7 is a gene which encodes a bone specific transcription factor required for osteoblast differentiation [Yoshida et al., 2012]. Mouse studies have demonstrated that osteoblast maturation arrest in Sp7-/- knockout mice result in bone formation failure [Nakashima et al., 2002; Yoshida et al., 2012]. In addition, a homozygous single base pair deletion (c.1052delA) in the SP7 gene has been reported in a child with osteogenesis imperfecta [Lapunzina et al., 2010]. The RARG gene encodes a retinoic acid receptor which is for a proximalizing factor that plays a morphogenetic role in pattern formation [Pennimpede et al., 2010]. Therefore, it is possible that SP7 and RARG play a role in skeletal anomalies observed in the previously reported patients. Furthermore, the severe neurological phenotype is not observed in the current family but is present in the reported patients. The largest 12q13.13-q13.2 deletion boundaries for the patient reported by Okamoto et al. [2011 contain at least 6 genes expressed in the central nervous system (GPR84, PDE1B, NPFF, SP1, ATP5G2, MAP3K12). The deletion in other two reported patients contains four of the six genes (NPFF, SP1, ATP5G2, MAP3K12) [Jonsson et al., 2012; Hancarova et al., 2013]. Since our patient with a mild neurological phenotype has the deletion containing only the two (GPR84 and PDE1B) of the six candidate genes for the neurological phenotype, the severe neurological phenotype in previously reported patients may be attributed by a contiguous deletion of multiple genes listed above. The variable neurological phenotype between the previously reported patients and the proband of the current family are the result of variable sizes of the deletion and spectrum of the genes involved in the deletion. The proband of current family has autism and developmental delay, while the sibling and mother have normal intelligence. The variable phenotype observed among the three members of our family may be explained by variable expressivity and incomplete penetrance of the deletion.
In summary, we report on a maternally inherited 12q13.13-q13.2 deletion in siblings with minor facial dysmorphic features but without evidence of skeletal anomalies. Although the proband has autistic-like features and mild developmental delay, the mother and daughter have normal intelligence. We hypothesize that the entire HOXC gene cluster deletion may be required for patients to manifest the abnormal skeletal phenotype. However, haploinsufficiency of additional genes found within the deletion boundaries may contribute to the severity of skeletal anomalies and intellectual disability. Variable expressivity and incomplete penetrance could contribute to the varying degree of clinical phenotypes, yet the possibility also exists that deletion of the HOXC gene cluster is not responsible for skeletal anomalies and intellectual disability.
ACKNOWLEDGMENTS
We thank the family for their cooperation and Dr. Areeg El-Gharbawy for her contribution in patient care.




