MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions†
How to Cite this Article: Stevens SJC, van Ravenswaaij-Arts CMA, Janssen JWH, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T, van Rheenen J, Heuts-Vijgen R, Stegmann APA, Smeets EEJGL, Engelen JJM. 2011. MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet Part A 155: 2739–2745.
Abstract
A partial deletion of chromosome band 2p25.3 (2pter) is a rarely described cytogenetic aberration in patients with intellectual disability (ID). Using microarrays we identified deletions of 2p25.3, sized 0.37–3.13 Mb, in three adult siblings and three unrelated patients. All patients had ID, obesity or overweight and/or a square-shaped stature without overt facial dysmorphic features. Combining our data with phenotypic and genotypic data of three patients from the literature we defined the minimal region of overlap which contained one gene, i.e., MYT1L. MYT1L is highly transcribed in the mouse embryonic brain where its expression is restricted to postmitotic differentiating neurons. In mouse-induced pluripotent stem cell (iPS) models, MYT1L is essential for inducing functional mature neurons. These resemble excitatory cortical neurons of the forebrain, suggesting a role for MYT1L in development of cognitive functions. Furthermore, MYT1L can directly convert human fibroblasts into functional neurons in conjunction with other transcription factors. MYT1L duplication was previously reported in schizophrenia, indicating that the gene is dosage-sensitive and that shared neurodevelopmental pathways may be affected in ID and schizophrenia. Finally, deletion of MYT1, another member of the Myelin Transcription Factor family involved in neurogenesis and highly similar to MYT1L, was recently described in ID as well. The identification of MYT1L as candidate gene for ID justifies further molecular studies aimed at detecting mutations and for mechanistic studies on its role in neuron development and on neuropathogenic effects of haploinsufficiency. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Nearly all hitherto reported deletions of chromosome band 2p25.3 (2pter) in patients with intellectual disability (ID) are accompanied by an inverted duplication of a proximal region of chromosome 2 [Bonaglia et al., 2009; Tirado et al., 2009]. The presence of both hemizygous and trisomic regions on the short arm of chromosome 2 in such cases hampers identification of candidate genes for ID and complicates genotype-phenotype correlations. Here we describe six patients with ID and pure terminal or interstitial deletions in 2p25.3 in which the segmental aneuploidies were characterized by SNP array or oligoarray-CGH. Using genotypic and phenotypic data from these six patients and from patients described in DECIPHER, we defined the minimal critical region of the 2p25.3 deletion phenotype. This report discusses the role of MYT1L as candidate gene for ID. MYT1L deletion gives a nonspecific clinical phenotype shared by patients with 2p25.3 deletions, with only ID and obesity/overweight being present in all patients.
METHODS
Cytogenetic and Molecular Genetic Analysis
Microscopic analysis of GTG-banded chromosomes was performed at a resolution of 550 bands per haploid genome using standard procedures. Fluorescent in situ hybridization (FISH) was done on metaphases as described by Lichter et al. [1988] using BAC-probe RP11-125K7 (kindly donated by dr. N. Carter), specific for chromosome band 2p25.3 (base pairs 607,981–753,912) and cosmid probe cCI2-578 specific for the centromere of chromosome 2.
The GeneChip® Human Mapping 250K NspI array (Affymetrix, Santa Clara, CA) was used for the analysis of DNA of the patients referred to the Maastricht University Medical Center (patients 1–4) using the manufacturer's protocol. This array has a mean spacing of approx. 1 probe per 10 kbp. Genome-wide copy number variations (CNVs) were determined by the Genotyping Console software version 3.0.2 (Affymetrix) using default software setting parameters. DNA samples of 40 healthy females were used as reference population. DNA of patients 5 and 6, referred to the University Medical Center Groningen, was analyzed using an Agilent 180k custom design oligonucleotide CGH-array (AMADID 23363 design), using the manufacturer's protocol (Agilent Technologies, Santa Clara, CA). Reference DNA consisted of a mixture of 40 female DNA samples. Data were processed using Feature Extraction V.9.1 and CGH analytics V. 3.4.27 (Agilent Technologies).
A copy number variant (CNV) was considered significant if four or more consecutive probes showed a gain or loss. A CNV was considered a normal genomic variant if the respective CNV had been detected in at least three control individuals as reported in the Database of Genomic Variants (http://projects.tcag.ca/variants) and/or encountered in at least three in-house control samples. Karyotypes were designated according to ISCN 2009 [Shaffer et al., 2009] and base pair positions were derived from the Genome Reference Consortium build GRCh37 (Ensembl release 57, March 2010).
CLINICAL REPORTS
Patients 1–3 are two brothers and a sister, aged 43, 52, and 57 years at diagnosis. They presented with moderate nonsyndromic ID, hyperactivity, mood changes and hypertension in absence of dysmorphic features. The sister is obese with a body mass index (BMI) of 32. Her height is 1.54 cm (−2.5 SD) and her weight is 75 kg (+3 SD) and she has a short, square-shaped stature. Both brothers are overweight with a BMI of 25 and 28, respectively. Their heights are 1.77 m (−1 SD) and 1.65 m (−2.5 SD) and their weights are 79 kg (+1 SD) and 76 kg (+2 SD), respectively (Fig. 1). All three patients were institutionalized and originate from a family with four other unaffected children and healthy, nonconsanguineous parents.
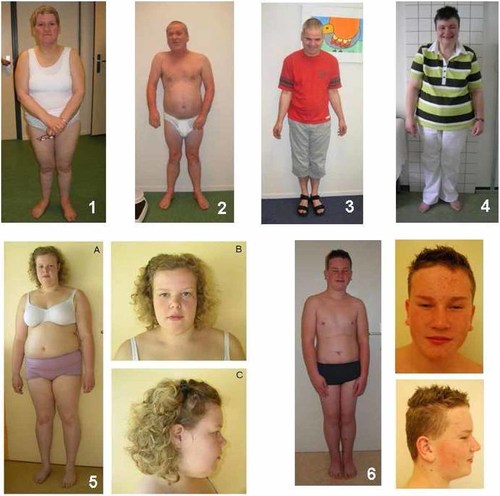
Photographs of patients 1–6 with intellectual disability and deletions in 2pter including the MYT1L gene. Note square truncal build in patients 1, 2, 4, and 5 with marked obesity in patients 1, 4, and 5 and overweight in the other patients.
Patient 4 is an unrelated, institutionalized 40-year-old female with moderate idiopathic ID and without dysmorphic features. She is obese (BMI = 36) with a short, square-shaped stature. Her height is 1.52 m (−3 SD) and her weight 82 kg (+3 SD; Fig. 1).
Patient 5 is the second child of healthy nonconsanguineous parents. Her psychomotor development was delayed with sitting unsupported at the age of 13 months, walking without support at 20 months and first words at the age of 2½ years. She experienced febrile convulsions at the age of 17 months and an EEG showed abnormal peak waves limited to the occipital regions. Incidentally a peak-wave complex was seen. She was treated by valproic acid for a period of three years, without any recurrence of convulsions. The girl developed obesity already during early childhood and she now has a square-shaped truncal build (BMI = 34). At the age of 15½ years her height was 161 cm (−1 SD), her weight was 88.5 kg (+3 SD) and her head circumference was 55 cm (0 SD). Apart from a broad forehead, mildly shortened palpebral fissures (ICD 50th centile, OCD 3rd centile), slightly upturned nose, and a short neck, there were no obvious facial dysmorphisms (Fig. 1). Neurological evaluation revealed mild hypotonia and hyperlaxity. Her IQ is 51 and she attends a school for children with moderate to severe learning problems. Behavioral problems were reported as aggressive outbursts in stressful situations.
Patient 6 is a 12-year-old boy with moderate ID. He was born after 35 + 6 weeks gestation with APGAR scores of 9 and 10 after 1 and 5 min. His birth weight was 2.785 kg (+0.3 SD) with height of 47.5 cm (−0.1 SD). He has had absences since his third year which are treated with Ethosuximide. He has a huge appetite and has been overweight since the age of 5. The first signs of puberty occurred at 7 years of age when pubic hairs appeared and penis and testes started to grow rapidly and acne developed. He was diagnosed with autistic spectrum disorder and attends a school for children with severe learning disabilities. He has a short attention span and is hypersensitive to hard noises. He gets tired easily, with subsequent exoforia. His gross and fine motor abilities are poor. Physical examination at 11.4 years revealed a head circumference of 54 cm (0 SD), length of 1.63 m (+1.9 SD), weight of 65 kg (weight for height +2 SD), ICD of 3.5 cm (>+2 SD), OCD of 8.2 cm (p25), palpebral fissure length of 2.5 cm (<<−2 SD) and ear length of 7.5 cm (>>+2 SD). Dysmorphic features included flat occiput, mild brachycephaly, large ears, short palpebral fissures with upslant and divergent strabismus. Heart auscultation revealed no murmur. Prominent abdomen with thick panniculus adiposus was noted. Tanner stage was PH4.
Routine karyotyping and DNA analysis for fragile X syndrome was performed in patients 1–6 and revealed no abnormalities.
RESULTS
Characterization of 2p25.3 Deletions by Microarray Analyses
SNP array analyses of DNA of the three siblings (Patients 1–3) showed a microscopally cryptic terminal deletion of chromosome band 2p25.3 and their karyotype was arr 2p25.3(1–2,769,875)x1. The hemizygous region was approx. 2.77 Mb in size and covered by 286 SNP array probes (SNP_A-1820282–SNP_A-2254308) (Fig. 2A). The deletion was confirmed by FISH using BAC probe RP11-125K7, which showed a fluorescent signal on the normal chromosome 2 only. It contained 10 transcribed genes: FAM110C, SHY3YL1, ACP1, FAM150B, TMEM18, C2ORF90, SNTG2, TPO, PXDN, and MYT1L. The four other, healthy siblings in this family were tested by metaphase FISH using probe RP11-125K7 and no aberrations were found. The parents of this family could not be investigated as they were deceased.
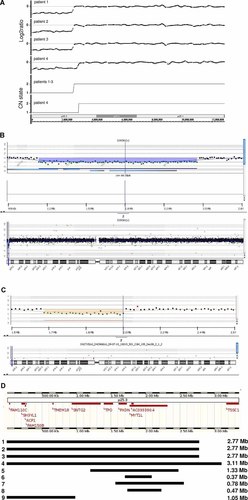
A: SNP array copy number analyses results for patients 1–4. Upper part shows log2 ratio of patients' versus references' signals, bottom shows the copy number state derived thereof. Patients 1–3 had a 2.77 Mb terminal deletion of chromosome band 2p25.3 and patient 4 a 3.11 Mb terminal deletion of this chromosome band. B: Oligonucleotide array-CGH results for patient 5 showing a loss of 1.33 Mb, i.e., an interstitial deletion in band 2p25.3. Upper part shows patient versus reference signal (76 oligonucleotides with decreased signal), bottom part shows the whole chromosome view. C: Oligonucleotide array-CGH results for patient 6 showing a loss of 0.37 Mb, i.e., an interstitial deletion in band 2p25.3. Upper part shows patient versus reference signal (24 oligonucleotides with decreased signal), bottom part shows the whole chromosome view. D: Schematic representation of deletions in chromosome band 2p25.3 in patients with intellectual disability (No. 1–8) and one patient without intellectual disability (No. 9). No. 1–3: patients 1–3 (siblings); No. 4: patient 4; No. 5: patient 5; No.6: patient 6; No. 7: DECIPHER case 141; No.8: DECIPHER case 255713; No. 9: DECIPHER case 249398. Note that deletion of MYT1L is shared between all patients with ID.
SNP array analysis of DNA of Patient 4 identified a similar de novo terminal deletion of part of chromosome band 2p25.3 and her karyotype was 46,XX.arr 2p25.3(1–3,127,370)x1dn. The hemizygous region was covered by 325 SNP array probes (SNP_A-1820282–SNP_A-1826128) and was approx. 3.13 Mb in size (Fig. 2A). The region contained the same 10 genes as in the three siblings described above.
Oligonucleotide CGH-array analysis of DNA of Patient 5 revealed a de novo loss comprising 76 oligonucleotides on the array specific for chromosome band 2p25.3. Her karyotype was 46,XX.arr 2p25.3(1,207,627–2,499,519)x1dn. The size of the interstitial deletion in 2p25.3 was approx. 1.33 Mb (Fig. 2B), with the last normal telomeric oligonucleotide at 1,191,676 bp (A_16_P35549183), the first oligonucleotide with a diminished signal at 1,207,627 bp (A_16_P35549209), the last oligonucleotide with a diminished signal at 2,499,519 bp (A_16_P15531589) and the first normal proximal oligonucleotide at 2,522,885 bp (A_16_P00301255). The region contained four genes: SNTG2, TPO, PXDN, and MYT1L.
Oligonucleotide CGH-array analysis of DNA of Patient 6 revealed a de novo interstitial deletion of approx. 0.37 Mb in band 2p25.3, designated as 46,XY.arr 2p25.3 (1711599–2078357)x1. The deletion comprised 24 oligonucleotides specific for 2p25.3 on the array. The last normal telomeric oligonucleotide was at 1,696,100 bp (A_16_P00299828), the first oligonucleotide with a diminished signal at 1,711,599 bp (A_16_P00299854), the last oligonucleotide with a diminished signal at 2,078,357 bp (A_18_P13083532) and the first normal proximal oligonucleotide at 2,092,651 bp (A_16_P35551726). The hemizygous region contains two genes: PXDN and part of MYT1L. Additionally, a 645 kb copy number gain in 5q35.1 was found, which was also seen in his normal father and which was interpreted as a familial variant.
Definition of the Smallest Region of Overlap of 2p25.3 Deletions in Patients With ID
We queried the DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) [Firth et al., 2009] for deletions in chromosome band 2p25.3 overlapping with those in the six patients described above. Five such cases were identified by array-based methods (patient identification numbers 00249399, 00248530, 00249398 and 00000141). Case 00249399 was excluded for further analysis because this was not a case of pure deletion 2p25.3. Case 00248530 was excluded because this probably concerned a benign copy number variant (CNV) inherited from a normal mother, comprising the FAM110C and SH3YL1 genes. It was located in a region with known benign loss CNVs (Database of Genomic Variants variations 3341, 50561, and 8936). The three remaining cases were a patient with ID, strabismus, tall stature, large hands and feet, obesity and short palpebral fissures (case 00000141), a patient with ID and autistic behavior (case 255713) and a patient suffering from psychotic behavior and hyperactivity but no ID (case 00249398). They showed an 0.78 Mb interstitial deletion affecting PXDN and MYT1L (case 00000141), an 0.47 Mb interstitial deletion affecting only MYT1L (case 255713) and a 1.05 Mb terminal deletion in band 2p25.3 affecting FAM110C, SHY3YL1, ACP1, FAM150B, TMEM18, and C2ORF90 (case 00249398), respectively (Fig. 2D, Patients 7–9). Subsequently, the smallest region of overlap of the deletions in the eight patients with ID (Patients 1–8 in Fig. 2D) could be defined as the region containing only MYT1L (myelin transcription factor 1-like), which is also known as NZF-1.
DISCUSSION
By combining data from three familial cases, three sporadic cases and three cases described in DECIPHER, we defined the shortest region of overlap for pure 2p25.3 deletions. These deletions are associated with ID and obesity/overweight (Fig. 2C). This region contains 1 protein-coding gene, i.e., MYT1L. Here we propose MYT1L haploinsufficiency as cause for ID.
MYT1L (OMIM 613084) encodes a neural transcription factor that has six characteristic zinc finger domains with CX4CX4HX7HX5C (CCHHC) consensus DNA binding sequence (Fig. 3) [Kim and Hudson, 1992; Kim et al., 1997; Romm et al., 2005]. In mouse and rat embryos it is solely transcribed in the developing central nervous system where it has a crucial role in neurogenesis [Jiang et al., 1996; Kim et al., 1997; Weiner and Chun, 1997]. Myt1L is expressed in a temporal manner during rodent embryogenesis, predominantly in differentiating, postmitotic neurons, including those in the cerebral cortex, thalamus, hindbrain, and dorsal root ganglia [Kim et al., 1997].
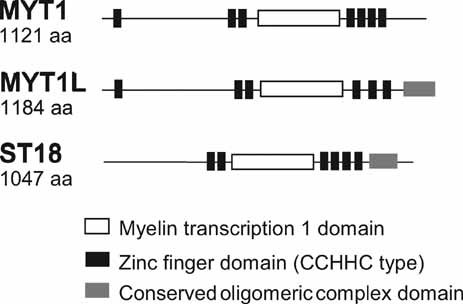
Schematic representation of conserved functional domains in the three members of the human myelin transcription factor protein family. Figure is based on HomoloGene Homo Sapiens entries 3332, 7435, and 8792, respectively (http://www.ncbi.nlm.nih.gov/homologene).
The pivotal role of MYT1L in regulating neuron differentiation is further underlined by a recent breakthrough study showing that the protein, in conjunction with transcription factor brn2, is essential for inducing functional mature neurons in a mouse induced pluripotent stem cell model [Vierbuchen et al., 2010]. Interestingly, these Myt1L-induced neurons resemble excitatory cortical neurons of the forebrain [Nicholas and Kriegstein, 2010; Vierbuchen et al., 2010], suggesting an important role for MYT1L in development of cognitive functions. Moreover, Myt1L, in conjunction with Ascl1, Brn2, and NeuroD1, can directly convert human fibroblasts into neural lineages [Pang et al., 2011]. These induced human neurons show neuronal morphology and express multiple neuronal markers [Pang et al., 2011; Pfisterer et al., 2011]. They are able to generate action potentials and to receive synaptic contacts and can be directed towards distinct functional neurotransmitter phenotypes, such as dopaminergic neurons [Pfisterer et al., 2011].
Unsurprisingly, MYT1L has been highly conserved during evolution. The human protein is 95% and 92% identical to mouse and rat Myt1l, respectively, with even higher identity in the zinc finger domains [Kim and Hudson, 1992]. MYT1L is a member of the myelin transcription factor protein family, which has two other members in humans, i.e., MYT1 and ST18 (Fig. 3). Deletion of MYT1 (OMIM 600379) was previously associated with ID as well [Kroepfl et al., 2008]. Like MYT1L, MYT1 is expressed exclusively in differentiating neurons in the developing embryonic nervous system [Kim and Hudson, 1992; Kim et al., 1997; Bellefroid et al., 1996]. MYT1 and MYT1L are highly related and show 62% identity at the protein level, similar functional domain topology (Fig. 3) and similar tissue-specific expression [Kim and Hudson, 1992; Kim et al., 1997]. Moreover, both proteins recruit the same histone deacetylases to regulate neural transcription via their interaction with Sin3B [Romm et al., 2005] pointing to similar regulatory functions in neurogenesis.
Duplication of MYT1L was recently described in schizophrenia patients, while in more than 700 controls no copy number variants comprising MYT1L were found [Vrijenhoek et al., 2008]. This indicates that MYT1L is a dosage-sensitive gene. Possibly, overlapping neurodevelopmental pathways are affected by MYT1L-related dosage effects in ID and other psychopathologies. This fits with emerging observations that recurrent pathogenic gains and losses are not disease-specific, but may be shared in for example ID, schizophrenia, and autism [Vrijenhoek et al., 2008; Guilmatre et al., 2009; O'Donovan et al., 2009]. The challenge for future investigations will be to define whether solely increased or decreased gene dosage during development underlies the psychopathological outcome or whether additional genetic or developmental factors are involved.
The patients described here do not exhibit overt facial dysmorphic features, but a short, square-shaped truncal build was apparent in 4/6 patients, with overweight/obesity being present in all six. Obesity was also reported in one of the two DECIPHER cases with MYT1L deletion. However, based on current literature, we could not find a direct association between MYT1L haploinsufficiency and overweight. Mouse knock-out models may be worthwhile to investigate whether MYT1L haploinsufficiency is directly causing the obesity phenotype or whether other genes on 2pter are involved. Interestingly, the obesity-associated TMEM18 gene is located in band 2p25.3 [Willer et al., 2009; Almén et al., 2010], and is deleted in 4 of our patients (Fig. 2D). It maps outside of the deletions in the other patients and studies investigating possible positional effects on TMEM18 expression may therefore be relevant.
Our findings justify molecular genetic studies aiming at detection of mutations in MYT1L and other members of the myelin transcription factor protein family. Furthermore, mechanistic studies on the role of MYT1L in neuron development and on neuropathogenic effects of haploinsufficiency are relevant to unravel pathways in which the protein is involved and its target genes.




