Complex distal 10q rearrangement in a girl with mild intellectual disability: Follow up of the patient and review of the literature of non-acrocentric satellited chromosomes†
How to Cite this Article: Sarri C, Douzgou S, Gyftodimou Y, Tümer Z, Ravn K, Pasparaki A, Sarafidou T, Kontos H, Kokotas H, Karadima G, Grigoriadou M, Pandelia E, Theodorou V, Moschonas NK, Petersen MB. 2011. Complex distal 10q rearrangement in a girl with mild intellectual disability: Follow up of the patient and review of the literature of non-acrocentric satellited chromosomes. Am J Med Genet Part A 155: 2841–2854.
Abstract
We report on an intellectually disabled girl with a de novo satellited chromosome 10 (10qs) and performed a review of the literature of the non-acrocentric satellited chromosomes (NASC). Satellites and stalks normally occur on the short arms of acrocentric chromosomes; however, the literature cites several reports of satellited non-acrocentric chromosomes, which presumably result from a translocation with an acrocentric chromosome. This is, to our knowledge, the third report of a 10qs chromosome. The phenotype observed in the proband prompted a search for a structural rearrangement of chromosome 10q. By microsatellite analysis we observed a 4 Mb deletion on the long arm of chromosome 10, approximately 145 kb from the telomere. FISH and array CGH analyses revealed a complex rearrangement involving in range from the centromere to the telomere: A 9.64 Mb 10q26.11–q26.2 duplication, a 1.3 Mb region with no copy number change, followed by a 5.62 Mb 10q26.2–q26.3 deletion and a translocation of satellite material. The homology between the repeat sequences at 10q subtelomere region and the sequences on the acrocentric short arms may explain the origin of the rearrangement and it is likely that the submicroscopic microdeletion and microduplication are responsible for the abnormal phenotype in our patient. The patient presented here, with a 15-year follow-up, manifests a distinct phenotype different from the 10q26 pure distal monosomy and trisomy syndromes. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
In humans, the nucleolus organizer regions (NORs) are usually located in the satellite stalks (secondary constrictions) in the short arms of the acrocentric chromosomes 13, 14, 15, 21, and 22. In situ hybridization studies have shown these sites to contain the 18S and 28S rRNA genes [Evans et al., 1974; Hsu et al., 1975]. The transcriptional activity of these genes can be shown by specific silver staining of the NORs [Miller et al., 1976].
Localization of NORs in positions other than the short arms of acrocentric chromosomes (ectopic NORs) are rare findings. They are thought to arise from translocations between acrocentric and non-acrocentric chromosomes or by insertion of a nucleolus organizer into a non-acrocentric chromosome. Depending on the particular rearrangement, carriers may or may not show phenotypic abnormalities.
Several reports of satellited non-acrocentric chromosomes exist in the literature (Table I). These unique chromosomes presumably result secondary to a translocation with an acrocentric chromosome. Generally, balanced carriers of a translocation are unaffected, but individuals with unbalanced reararrangements manifest anomalies because of partial trisomy, monosomy, or both.
| Satellited chromosome | Acrocentric chromosome involved | Inheritance | Abnormal phenotypea | Reference |
|---|---|---|---|---|
| 1q11 | 13 | De novo | Unknown | Mikkelsen et al. 1980 |
| 1q11 | 13 | Familial | None (B) | Lucas et al. 1972 |
| 1qter | D | Unknown | Unknown | Arai et al. 1994 |
| 1pter | 21 | De novo | Yes (U) | Ki et al. 2003 |
| 1p | 15 | Familial | Yes (U) | Barbi et al. 1992 |
| 1pter | 15 | De novo | Yes (U) | Habibian et al. 1994 |
| 1pter | Unknown | Familial | None (U) | Habibian et al. 1994 |
| 1q | 13 | Familial (?) | None | Lucas et al. 1972 |
| 1q44-qter | 15 | Familial | Yes (U) | Villa et al. 1998 |
| 2pter | Unknown | Familial | None (U) | Elliott and Barnes 1992 |
| 2qter | Unknown | De novo | Yes (U) | Weise et al. 2002 |
| 2qter | Unknown | Familial | None (U) | Bauld and Ellis 1984 |
| 2qter (6 cases) | Unknown | Familial (5 cases) | None (?) | Lamb et al. 1995 |
| Unknown (1 case) | ||||
| 2qter | Unknown | De novo | None (B) | Reddy and Sulcova 1998 |
| 2qter | Unknown | Familial | Unknown | Bauld and Ellis 1984 |
| 3q | 15 | Familial | None (B) | Hansmann et al. 1977 |
| 4qter | Unknown | Familial | None (U) | Babu et al. 1987 |
| 4q35 | Unknown | De novob | Yes (U) | Babu et al. 1987 |
| 4q35 | Unknown | Familial | None (U) | Mihelick et al. 1984 |
| 4q35 | Unknown | Familial | Yes (U) | Mihelick et al. 1984 |
| 4q35 | Unknown | Familial | None (U) | Hawks-Arn et al. 1995 |
| 4p16.3 | 21 | Familial | None (B) | Hawks-Arn et al. 1995 |
| 4q35 | Unknown | Unknown | None (U) | Hawks-Arn et al. 1995 |
| 4p16.3 | Unknown | Familial | Yes (U) | Estabrooks et al. 1992 |
| 4p16 | 15p11.1 | Unknown | None (B) | Chen et al. 2000 |
| 4qs | 21 | Familial | None (B) | Corti et al. 1999 |
| 4q21 | 14p12 | Familial | None (B) | Grabowski et al. 2000 |
| 4qter | Unknown | Familial | None (U) | Guttenbach et al. 1999 |
| 4qter | Unknown | Familial | None (U) | Miller et al. 1995 |
| 4p16.3 | 15 | De novo | Yes | Iconen et al. 1992 |
| 4qter | 14 or 22 | Familial | None (U) | Shah et al. 1997 |
| 4ps | Unknown | Unknown | Yes (U) | Our case unpublished |
| 5p | 13 | Familial | Yes (U) | Dev et al. 1979 |
| 6p | 21 | Familial | Infertility (B) | Dahoun et al. 1997 |
| 6q15 | Unknown | Familial | Yes (U) | Prieto et al. 1989 |
| 7q21.3-q22.1 | Unknown | Familial | None(U) | Guttenbach et al. 1998 |
| 7q11.3 | 13p13 | Unknown | Unknown | Mikkelsen et al. 1980 |
| 7qs | 22 | De novo | Yes (U) | Corti et al. 1999 |
| 8q11 | Unknown | Familial | None(U) | Guttenbach et al. 1999 |
| 9p11 | 13 | Familial | None (B) | Varley et al. 1981 |
| 9q | 13 | Familial | None (B) | Hansmann et al. 1977 |
| 9q34.3 | 22 | Familial | Yes (B) | Harada et al. 1989 |
| 10p13 | 13p12 | Familial | Yes (B) | Faivre et al. 1999 |
| 10qter | Unknown | Familial | Normal (U) | Storto et al. 1999 |
| 10qs | 13 | Familial | Yes | Lese et al. 1998 |
| 11 | 22 | Familial | Tomkins 1981 | |
| 11q21 | Unknown | Familial | Unknown | Cosper et al. 1985 |
| 12ps (4 cases) | Unknown | Familial | None (6 cases) Short stature (1 case) | Willatt et al. 2001 |
| 12p | Unknown | Familial | None (B) | Watt et al. 1984 |
| 12p11 | 21 | Familial | None (B) | Parslow et al. 1979 |
| 12q24 | 21p11 | Familial | Unknown | Mikkelsen et al. 1980 |
| 15qs | Unknown | De novo | Yes | Rujirabanjerd et al. 2007 |
| 16q23 | 13 | Familial | Yes (U) | Savary et al. 1991 |
| 17ps | ? | Familial | Unknown | Killos et al. 1997 |
| 17ps | 15 | De novo | Unknown | Lese et al. 1998 |
| 18p | 13 | Familial | None (B) | Van Tuinen et al. 1983 |
| 21q22.1 | Unknown | De novo | Yes | Chen et al. 2004 |
| Xp | 21 | De novo | Yes (U) | Stetten et al. 1986 |
| Xp21 | 21 | De novo | Yes (B) | Verellen-Dumoulin et al. 1984 |
| Xqter | Unknown | Familial | Yes (U) | Chen et al. 2000 |
| Yqs (3 cases) | Unknown | Familial | None (B) | Review in Schmid et al. 1984 |
| Yqs (2 cases) | Unknown | Unknown | None (U) | Chandley et al. 1989 |
| Yqs | Unknown | Familial | None (U) | Genest, 1973 |
| Yqs | Unknown | Familial | None (U) | Genest, 1978 |
| Yqs | Unknown | Familial | None (U) | Genest, 1979 |
| Yqs | 14 | De novo | None (U) | Genest et al. 1983 |
| Yps | Unknown | De novo | None (U) | Lin et al. 1995 |
| Yqs | Unknown | Familial | Yes | Martin et al. 1982 |
| Yps | 15 | De novo | None (B) | Reddy, 1998 |
| Yqs | 15 | Familial (B) | None | Verma et al. 1997 |
| Yqs (3 cases) | 15 | Familial (B) | Nome | Wilkinson and Crolla 1993 |
| Yqs | Unknown | Familial | None | Shabtai et al. 1981 |
| Yqs | 15 | De novo | None | Velissariou et al. 2001 |
| Yqs (4 cases) | 15 (2 cases) the rest unknown | Familial | None | Kühl et al. 2001 |
| Yqs (2 cases) | 15 | Familial | None | Velissariou et al. 2007 |
- Note: A list of previously reported non-acrocentric satellited chromosomes is given along with the acrocentric chromosome involved in the translocation, the inheritance pattern, and the phenotypic status of balanced and unbalanced carriers.
- a U, unbalanced translocation carrier with only the satellited chromosome present in the karyotype; B, balanced translocation carrier.
- b Affected patient had a de novo interstitial deletion.
Males with a Yqs chromosome are generally not affected by the deletion of Yq12, because this region of the Y does not contain expressed genes.
There are several additional reports of satellited non-acrocentrics (1p, 1q, 2p, 2q, 3q, 4p, 4q, 9p, 9q, 10q, 12p, 18p, Yp, Yq) in which the carriers are not affected (Table I). In these cases, the satellites may have been re-inserted in the telomeric regions so that no phenotypically important genetic material was lost.
In other reports the carriers of a satellited non-acrocentric chromosome are adversely affected because of proven or possible loss of genetic material. Such cases have been described regarding chromosomes 1p, [Barbi et al., 1992; Habibian et al., 1994; Ki et al., 2003], 1q [Villa et al., 1998], 2q [Weise et al., 2002], 4p [Estabrooks et al., 1992; Iconen et al., 1992], 4q [Mihelick et al., 1984; Babu et al., 1987], 5p [Dev et al., 1979], 7q [Corti et al., 1999], 9q [Harada et al., 1989], 10p [Faivre et al., 1999], 10q [Lese et al., 1998], 15q [Rujirabanjerd et al., 2007], 21q [Chen et al., 2004], Xp [Stetten et al., 1986; Verellen-Dumoulin et al., 1984], Xq [Chen et al., 2000], and one case of Yq [Martin et al., 1982]. There are a few other reports of satellited non-acrocentric chromosomes with unspecified phenotypes involving chromosomes 1q [Mikkelsen et al., 1980; Arai et al., 1994], 2q [Bauld and Ellis, 1984; Lamb et al., 1995], 7q [Mikkelsen et al., 1980], 12q [Mikkelsen et al., 1980], and 17p [Killos et al., 1997; Lese et al., 1998].
We have identified a 10-month-old girl with a de novo satellited chromosome 10q (10qs). The proband carries the 10qs chromosome without the reciprocal derivative acrocentric and should have both a duplication of the NOR, which presumably would not have a phenotypic effect, as well as a microdeletion and a microduplication of some portion of chromosome 10q26.
Reports of 10q terminal deletions without a familial translocation or other chromosomal abnormality are uncommon, and few cases with breakpoints in 10q26 have been reported so far. Interstitial deletions of chromosome 10 involving the same region are even less frequent.
The chromosomal breakpoint for most of the reported 10q terminal deletion cases resides at 10q26 [Turleau et al., 1979; Taysi et al., 1982; Evans-Jones et al., 1983; Zatterale et al., 1983; Shapiro et al., 1985; Mehta et al., 1987; Fryns et al., 1989; Greenberg et al., 1989; Gorinati et al., 1989; Wulfsberg et al., 1989; Kogasaka et al., 1990; Schrander-Stumpel et al., 1991; Teyssier et al., 1992; Wilkie et al., 1993; Petit et al., 1998; Leonard et al., 1999; Ogata et al., 2000; Lucusa et al., 2002; Wu et al., 2002; Scigliano et al., 2004].
In this study, we describe to our knowledge the first 10qs case reported with a molecularly verified deletion/duplication and distinct phenotype and we present FISH, molecular and array CGH analysis of the involved chromosomal region. We also review the literature on clinically similar cases.
CLINICAL REPORT
A 10-month-old girl was referred for clinical evaluation because of dysmorphism. She was the second child of healthy non-consanguineous Cretan parents. An older brother was clinically normal. At birth, the mother was 31 years old and the father 34. Family history was unremarkable. Pregnancy was complicated by a 5 day long bleeding episode during the first trimester, for which bed rest was advised. The mother smoked eight to 10 cigarettes per day. Serologic tests for cytomegalovirus, toxoplasmosis, and rubella, and triple serum marker screening were negative. A standard ultrasound scan at 22½ weeks of gestation was normal. The patient was born at 38 weeks gestation by cesarean due to placental detachment. Birth weight was 2.060 g (<3rd centile), length 45 cm (3rd centile), and OFC 32.5 cm (10th centile). She presented with left clubfoot. Apgar scores were not recorded. She received oxygen for 24 hr and remained in incubator for 7 days. Cord blood gas values were normal, no resuscitation was required and pediatric monitoring did not reveal any end-organ dysfunction. Brain ultrasound and magnetic resonance imaging, spine radiographs, heart and abdominal ultrasound as well as standard biochemical testing were unremarkable. Auditory brainstem responses and evaluation of vision and fundus were normal. At the age of 6 months, clubfoot was surgically corrected.
On physical examination at the age of 10 months she had a narrow face with bitemporal narrowing, convergent eyebrows, bilateral epicanthal folds, and long eyelashes. She presented with marked prognathism, short neck, and convergent strabismus (Fig. 1). Oral inspection disclosed a highly arched palate and gingival overgrowth. Enlarged gingiva was noted that was normal in color and firm on palpation. She was hypotonic and could not sit unsupported. On clinical evaluation, both parents were normal.
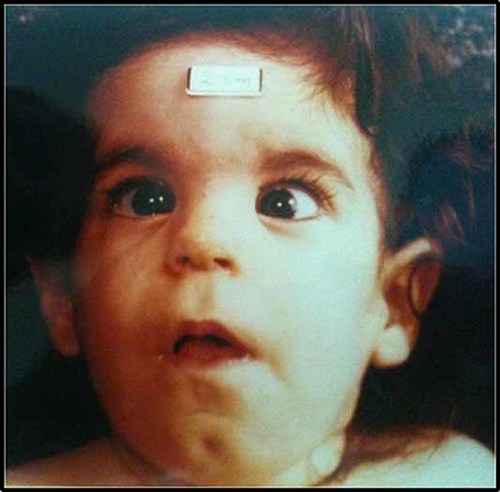
Patient at the age of 10 months. Note bilateral epicanthus, broad nasal bridge, strabismus.
The patient was lost to follow-up until the age of 15 years (Fig. 2). On re-evaluation she was 1.55 m tall (15th centile) with an OFC of 57.5 cm (95th centile). Her face was quite coarse, long, and asymmetric with prominence of the right side. Multiple naevi were present on the prominent hemiface. She had deep-set eyes, a broad nasal root with prominent nasal bridge, small nares, prognathism, and a short neck (Fig. 2A,B,C). Although it had not been possible to perform X-rays, peculiar hands and feet were apparent. Hands were long but brachytelephalangic with nail hypoplasia. This feature was mostly evident on the thumb. A bilateral sandal gap was evident on both feet, toes appeared short but nails were normal (Fig. 2D,E).
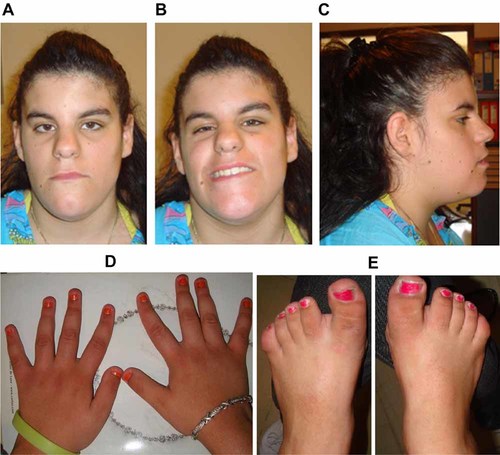
Patient at the age of 15 years. A: Long triangular face. B: Facial asymmetry, prominent on smiling. C: Multiple facial naevi on the prominent hemiface. D: Thin fingers with apparent brachytelephalangy, nail hypoplasia, and broad thumbs. E: Apparent brachydactyly, broad halluces, and sandal gap.
Developmental milestones were reportedly delayed: She sat at the age of 11 months, walked at age of 2 years, and acquired language skills at the age of 18 months. The patient had mild intellectual disability. She participated in many daily activities, attended a special school, and had very good social skills. The father reported two brief psychotic episodes in the patient that followed parents' divorce. One of them ended with a suicidal attempt that prompted treatment with risperidone. The treatment was interrupted after 2 months because of adverse drug effects, mainly sedation, and it was not followed by further episodes. Metabolic work up including serum lactate, ammonia, amino acids, organic acids, cholesterol, very long chain fatty acids, lysosomal enzymes, transferrin isoelectric focusing, and urinary mucopolysaccharides were normal. Sleep electroencephalography recording was normal.
MATERIALS AND METHODS
Standard cytogenetic GTG, CBG, and AgNOR banding studies were performed on blood lymphocytes.
Probes for FISH were prepared and used according to standard protocols [Pinkel et al., 1986]. The probes used in this study were telomere specific DNA probes (All Human Telomeres, Oncor), β-satellite DNA probe specific for chromosome 10 centromere (D10Z1, Oncor), whole chromosome 10 paint probe (Cambio), 10q subtelomere probe (TL1002, Li Star FISH), and 28S rRNA probe and YAC (Yeast Artificial Chromosome) probes.
For the molecular analysis, 25 short sequence repeat (SSR, microsatellite) DNA polymorphisms on distal 10q were used (Table II) [Dib et al., 1996; Rosenberg et al., 1997].
| Locus | Marker map positiona (bps from 10pter) | Paternal genotype | Maternal genotype | Proband genotype | Conclusion |
|---|---|---|---|---|---|
| D10S579 | 89430125–89430392 | 1,2 | 1,1 | 1,2 | normal |
| D10S192 | 102436227–102436461 | 1,2 | 2,3 | 1,3 | normal |
| D10S597 | 111230789–111231008 | 1,2 | 1,1 | 1,2 | normal |
| D10S562 | 116640361–116640545 | 2,3 | 1,4 | 1,3 | normal |
| D10S1156 | 118332601–118332930 | 2,3 | 1,3 | 1,3 | normal |
| D10S187 | 118653033–118653260 | 1,2 | 2,3 | 2,- or 2,2 | ni |
| D10S587 | 125188640–125188789 | 2,3 | 1,3 | 1,3 | normal |
| D10S1213 | 125406637–125406748 | 1,1 | 1,2 | 1,2 | normal |
| D10S186 | 128790910–128791124 | 3,4 | 1,2 | 1,3 | normal |
| D10S1782 | 129086934–129087196 | 1,2 | 2,2 | 1,2 | normal |
| D10S1222 | 129260749–129260972 | 1,2 | 1,1 | 1,2 | normal |
| D10S1727 | 129326646–129326916 | 1,2 | 1,1 | 1,- or 1,1 | ni |
| D10S217 | 129540247–129540423 | 2,2 | 1,3 | 3,- | pat del |
| D10S1676 | 130006232–130006395 | 1,1 | 1,2 | 2,- | pat del |
| D10S1439 | 130593139–130593323 | 2,2 | 1,1 | 1,- | pat del |
| D10S1655 | 130955935–130956182 | 1,1 | 1,1 | 1,- or 1,1 | ni |
| D10S505 | 131915365–131915611 | 2,3 | 1,2 | 2,- or 2,2 | ni |
| D10S169 | 132521626–132521738 | 2,3 | 2,2 | 2,- or 2,2 | ni |
| D10S1770 | 132685760–132685965 | 1,3 | 2,2 | 2,- | pat del |
| D10S1651 | 132692559–132692754 | 2,3 | 1,1 | 1,- | pat del |
| D10S555 | 133443642–133443773 | 1,1 | 1,1 | 1,- or 1,1 | ni |
| D10S1675 | 133578326–133578564 | 1,2 | 1,3 | 3,- | pat del |
| D10S590 | 133647596–133647840 | 1,2 | 1,1 | 1,- or 1,1 | ni |
| D10S212 | 134449601–134449789 | 2,2 | 1,2 | 2,- or 2,2 | ni |
| D10S1711 | 135203794–135203971 | 1,1 | 1,1 | 1,- or 1,1 | ni |
- Abbreviations: ni, non-informative; pat del, deletion of the paternal allele.
- a Ensembl Genome Browser (http://www.ensembl.org/).
The alleles were analyzed after polymerase chain reaction (PCR) amplification of genomic DNA with end-labeling of one primer with 32P, polyacrylamide gel electrophoresis of the amplification products, and autoradiography of the dried gels [Petersen et al., 1990].
Array CGH was carried out using CytoChip BAC array (version 2.0.1, BlueGnome Ltd., Cambridge, UK). The chromosome positions are given according to the human genome assembly GRCh37 (Hg19).
Quantitative PCR (qPCR) was carried out using SYBR Green I detection chemistry and the 7500 Fast Real-Time PCR System (Applied Biosystems).
Informed written consent for publishing the images of the proband was obtained from her father.
RESULTS
Using GTG-banding, the karyotype of the proband was established as 46,XX,10qs de novo (Fig. 3, left panel). The satellite on the 10q telomere was positive with AgNOR banding (Fig. 4), but negative with CBG banding. Similarly FISH analyses using 28S rRNA probe gave signal on the most telomeric part of 10qs, besides the short arms of all the acrocentric chromosomes, supporting that this region contained satellite sequences. Both parents had a normal karyotype.
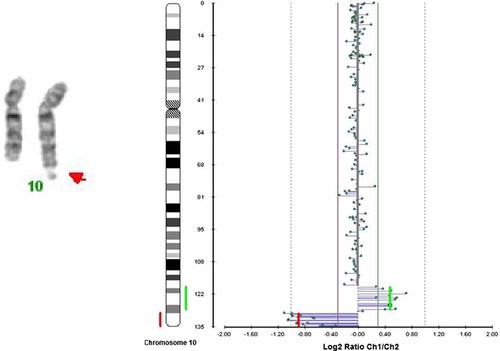
Left panel: Partial karyotype of the proband (GTG-banding). Arrow shows the satellited chromosome 10q. Right panel: Array CGH analysis of chromosome 10 of the proband showing a microdeletion in the 10q26.2-10q26.3 region (red line) and a microduplication in the 10q26.11-10q26.2 subtelomeric region (green line). Between the deletion/duplication region there is a small region of “no change.”
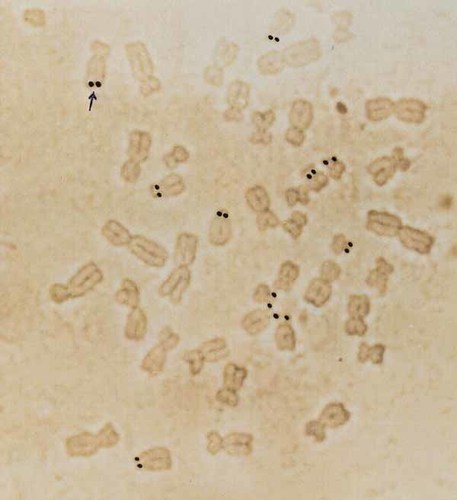
The satellited chromosome 10q, is positive with AgNOR banding (arrow) besides the also positive NOR regions of all the acrocentric chromosomes.
The whole chromosome 10 paint probe showed signals throughout both chromosomes 10, except for the satellite as expected. The 10q subtelomere probe displayed signals on both the normal and the satellited chromosome 10. The all-human telomere probe, apart from the telomeres of all the other chromosomes, gave two signals on the 10qs chromosome, one signal at the 10p telomere and the other on the satellite (data not shown). The 28S rRNA probe showed hybridization signals on all acrocentric chromosomes and on the 10qs satellites. Chromosome 10 centromere specific probes showed signals only on the centromeres of both chromosomes 10, but not on the satellite of 10qs. FISH analysis using YAC (Yeast Artificial Chromosome) probes, showed the presence of telomeres in the 10qs chromosome and a microduplication and a microdeletion in the long arm of the satellited 10q chromosome.
Molecular analysis was performed using 25 microsatellites from the 10q26-qter region (Table II). The polymorphic markers D10S217, D10S1676, D10S1439, D10S1770, D10S1651, and D10S1675 suggested an approximately 4 Mb deletion of paternal origin.
These results were confirmed with FISH analyses using YAC probes mapping to this region (data not shown). As FISH analyses also suggested a duplication proximal to the deleted region, we carried out array CGH to map the deletion and the duplication.
Array CGH results (Fig. 3, Right panel) suggested a 9.5–11 Mb interstitial duplication at 10q26.11–q26.2 (chromosome position of the minimum boundaries: chr 10:118,699,996–127,552,632 and maximum boundaries: chr 10:117,972,993–128,908,525). Approximately 1.3 Mb distal to the duplication an approximately 5.62 Mb interstitial deletion was present at 10q26.2–q26.3 (chromosome position of the minimum boundaries: chr 10:129,731,124–135,390,508; and maximum boundaries: chr 10:128,785,573–135,534,747) approximately 145 kb from the 10q telomere.
Both the deletion and duplication were verified with qPCR; investigation of the parents indicated that both events had occurred de novo. The duplication/deletion breakpoints were further mapped using qPCR. The proximal breakpoint of the duplication was mapped within an approximately 390 kb region (chromosome position chr 10: 118,228,711–118,616,009) and the distal breakpoint of the duplication was mapped within an approximately 300 kb region (chromosome position chr 10: 128,594,016–128,910,525). The deletion started within an approximately 270 kb region (chr 10: 129,076,897–129,350,817) and extended to the telomere. Between the proximal deletion and the distal duplication there was an approximately 1 Mb region which was intact.
DISCUSSION
Review of the Literature of Non-Acrocentric Satellited Chromosomes
Non-acrocentric satellited chromosomes with variable phenotypic effects
Unbalanced carriers of satellited autosomes may or may not display phenotypic abnormalities [Estabrooks et al., 1992; Iconen et al., 1992; Bauld and Ellis, 1984; Elliott and Barnes, 1992; Miller et al., 1995; Wang and Chen, 2004]. The phenotypic effects seen have usually been secondary to monosomy or trisomy for part of the chromosomes involved in the translocation.
The variable effects seen in some cases are not easily explained [Dahoun et al., 1997; Killos et al., 1997; Verma et al., 1997; Lese et al., 1998]. Individuals who carried a 4ps chromosome showed both normal and abnormal phenotypes [Estabrooks et al., 1992; Iconen et al., 1992; Hawks-Arn et al., 1995; Chen et al., 2000].
Babu et al. [1987] described a family where the father and five children had a 4q35s. The carrier father and four of the five children were phenotypically normal while one child had developmental delay and a possible de novo interstitial deletion of 4q35 on the satellited chromosome 4.
Estabrooks et al. [1992] described a normal father and his mother to be carriers of a 4p16.3s. His two children had developmental delay and the 4p16.3s.
Hawks-Arn et al. [1995] described three families with a satellited chromosome 4 with the following findings: (a) A normal carrier mother with recurrent pregnancy loss and a fetus with trisomy 2 and 4qs, and the maternal grandmother—also carrier of the 4qs—without any pregnancy loss. (b) Another family with a fetus with 47,XYY, 4qs karyotype who was phenotypically normal at birth, and (c) a third family with a fetus, the father, and the older child having a 4p16.3s. All of them were phenotypically normal and the mother had no miscarriages.
Dahoun et al. [1997] described a family with a 6ps chromosome. The mother and her five children (two girls and three boys) had the 6ps chromosome. The three boys were all infertile while the two girls and the mother were fertile.
Savary et al. [1991] described a female newborn with partial trisomy of the long arm of chromosome 16. The chromosome anomaly was the result of an unbalanced segregation of a balanced maternal translocation t(13;16)(p12;q23) where nucleolar organizers were present on the tip of the long arms of the derivative 16 maternal chromosome.
Non-acrocentric satellited chromosomes with abnormal phenotype
There are also cases where the phenotype was described as abnormal [Dev et al., 1979; Martin et al., 1982; Verellen-Dumoulin et al., 1984; Stetten et al., 1986; Babu et al., 1987; Harada et al., 1989; Barbi et al., 1992; Iconen et al., 1992; Habibian et al., 1994; Villa et al., 1998; Faivre et al., 1999; Weise et al., 2002].
Barbi et al. [1992] described a child with a chromosome 1p36.3s and a partial monosomy 1pter as a result of a balanced translocation t(1;15) (p36.3;ps) in his mother. The child had generalized muscular hypotonia and other minor anomalies.
Chen et al. [2004] described an infant with a de novo 21qs associated with corpus callosum dysgenesis, colpocephaly, a concealed penis, congenital heart defects, and developmental delay.
In our Department we have seen three cases with satellited non-acrocentric chromosomes (three in a total of 13,000 postnatal karyotypes or 0.02%). The one family had a little girl with mild psychomotor retardation, ectopia of the kidney, strabismus, and a de novo 2qs chromosome, while the second family had a child with psychomotor retardation, somatic delay, and a 4ps chromosome (unpublished data). The third case is the 10qs presented here.
Non-acrocentric satellited chromosomes with normal phenotype
In other cases the phenotype was reported normal [Lucas et al., 1972; Genest, 1973; Hansmann et al., 1977; Genest, 1978, 1979; Parslow et al., 1979; Varley et al., 1981; Genest et al., 1983; Van Tuinen et al., 1983; Bauld and Ellis, 1984; Mihelick et al., 1984; Schmid et al., 1984; Babu et al., 1987; Chandley et al., 1989; Elliott and Barnes, 1992; Wilkinson and Crolla, 1993; Hawks-Arn et al., 1995; Lin et al., 1995; O'Malley et al., 1997; Shah et al., 1997; Verma et al., 1997; Reddy, 1998; Reddy and Sulcova, 1998; Corti et al., 1999; Guttenbach et al., 1999; Storto et al., 1999; Grabowski et al., 2000; Kühl et al., 2001; Velissariou et al., 2001; Willatt et al., 2001].
Miller et al. [1995] described a male fetus with a 4qs chromosome transmitted through three generations without any obvious phenotypic effects.
Wilkinson and Crolla [1993] described three families with t(15;Y)(ps;qter) without any phenotypic abnormalities.
Interstitial insertions of NOR regions
Most authors describe translocations of a satellited region to the subterminal p or q end of a chromosome. Interstitial insertions of NORs into non-acrocentric chromosomes are observed even more rarely. Although such rearrangements have been described in some tumor cell lines [Neerman-Arbez et al., 1993; Atkin and Baker, 1995], only 12 non-tumor cases have been reported to our knowledge.
Most of them are familial with normal [Parslow et al., 1979; Varley et al., 1981; Watt et al., 1984; Guttenbach et al., 1998, 1999; Grabowski et al., 2000], or abnormal [Prieto et al., 1989; Faivre et al., 1999] phenotype. One case referring to chromosome 7q11.3 is of unknown inheritance and phenotype [Mikkelsen et al., 1980].
A rearranged familial insertion of NOR in chromosome 11q21 was detected at prenatal diagnosis with unknown outcome. In this report two active NORs joined by a centromere had been inserted into 11q21 [Cosper et al., 1985]. Two de novo cases with abnormal outcomes have been reported so far regarding insertions in chromosome 21q22.1 [Chen et al., 2004], and chromosome Xp21 [Verellen-Dumoulin et al., 1984], respectively.
The satellited Y chromosomes
Satellited non-acrocentric chromosomes are rare events. The chromosome most often involved appears to be the Y with more than 30, mostly familial, reported cases of Yqs (for review see Schmid et al., 1984) and two cases of Yps [Lin et al., 1995; Reddy, 1998] (Table I).
It is generally accepted that the Yqs chromosomes are derived from reciprocal translocation events between a normal Y chromosome and an acrocentric chromosome. Thus, the structure of the Yqs chromosomes can show considerable interfamilial difference. In general, the presence of an additional NOR in the Y chromosome heterochromatin region does not cause any phenotypic abnormalities [Schmid et al., 1984].
Schmid et al. [1984] described 18 families with a Yqs chromosome. No phenotypic abnormality could be directly related to this chromosomal abnormality. Indirectly it could be hypothesized to be the cause of chromosome aneuploidies such as trisomies (Down or Klinefelter syndromes) or unbalanced translocations.
Previous detailed pedigree analysis [Genest, 1973, 1978; Genest et al., 1983] led to the surprising finding that a Yqs chromosome had been transmitted in paternal lineage for 315 years and was the oldest chromosomal translocation described in mankind. The Yqs chromosome was revealed after 11 generations of transmission from 16 males when at the eleventh generation one of the Yqs carriers gave birth to a child with Down syndrome.
Chandley et al. [1989] described a deleted Yq in the sterile son of a man with a satellited Yq chromosome. The other son of the family had the satellited Yqs and was fertile like the father.
Lin et al. [1995] described a phenotypically normal Yqs carrier father who did not transmit the satellite to his son, probably due to meiotic loss.
Kühl et al. [2001] described loss of the Y chromosomal PAR2-region in four familial cases of satellited Y chromosome (Yps) where the carriers of the satellited Y developed normally. Loss of the Y chromosome PAR2-region and additional rearrangements in two familial cases of satellited Y chromosome were also described by Velissariou et al. [2007].
Non-acrocentric satellited chromosomes in prenatal diagnosis
The presence of a non-acrocentric satellited chromosome in the karyotype of a fetus during prenatal diagnosis will often give rise to problematic genetic counseling. It has been speculated that carriers of NOR variants may be at risk of spontaneous abortions due to abnormal segregation, but this does not seem the case with double NORs [Hassold et al., 1987].
Mihelick et al. [1984] described a fetus with a 4q35s of paternal origin with acrania, anencephaly, craniorachischisis, and absent spinal cord. The carrier father was normal.
Killos et al. [1997] described a female fetus and her father having a 17ps chromosome, with a deletion outside the Miller–Dieker syndrome critical region. The 17ps chromosome was not considered as having any clinical consequence since the carrier father was normal, and the pregnancy was not interrupted.
Chen et al. [2000] described a family with female twin fetuses with a 4ps derived from a cryptic balanced translocation t(4;15)(p16;p11.1) of the mother who already had a child with Wolf–Hirschhorn syndrome. The same authors, in the same publication, described a male fetus with an Xqs of maternal origin. The mother was phenotypically normal while the male fetus, who had a deletion of the subtelomeric region of Xq, developed psychomotor retardation and other congenital abnormalities. The authors suggested that pregnant women with satellited non-acrocentrics are at risk of carrying fetuses with chromosomal abnormalities. If the X chromosome is involved, the male fetus can be affected by X-linked recessive disorders, including mental retardation.
Ki et al. [2003] described a fetus as intrauterine growth retarded, with an ectopic NOR on 1p and a ring chromosome 21. A female infant was delivered small for gestational age with numerous subtle physical abnormalities, neurologically normal.
Satellited 10q chromosomes
Concerning satellited chromosome 10q only two cases have been reported so far [O'Malley et al., 1997; Storto et al., 1999 (same case); Lese et al., 1998]. In the case of O'Malley et al. [1997] and Storto et al. [1999] (same case), a phenotypically normal female fetus inherited a 10qs chromosome from her phenotypically normal father who was mosaic for the 10qs. FISH with a 10q subtelomeric probe did not demonstrate deletion neither in amniocytes nor in paternal blood. In the second reported case [Lese et al., 1998] a 10qs chromosome of maternal origin was identified in an amniotic fluid sample. It was the derivative result of a maternal balanced 10q;13p translocation and the fetus was monosomic for distal 10q.
To our knowledge our case is the third reported case with 10qs and the first case studied with molecular analysis and array CGH methods.
Comparison to monosomy 10q26-qter and to 10q26-qter trisomy reported cases
Array CGH data showed an approximately 9.64 Mb interstitial microduplication at 10q26.11–q26.2 and an approximately 5.62 Mb interstitial microdeletion at 10q26.2–q26.3 region of satellited chromosome 10 (Fig. 3, right panel). The microsatellite analysis demonstrated the absence of at least six informative marker alleles but could not detect the microduplication. Array CGH and qPCR confirmed the FISH (YAC) data.
The patient reported here shares many features with the pure monosomy 10q26-qter patients (Table III). Her perinatal phenotype is consistent with most of the 10q26-qter features yet it can be distinguished by the absence of microcephaly. The facial morphology at 10 months and 15 years is reminiscent of the photographs reported by Lucusa and Fryns [2000] and by Leonard et al. [1999]. The distinctive facial asymmetry, long triangular face, and broad nasal bridge are present also in the present patient, with the 10q26.2–10q26.3 band deletion, suggesting that dosage-sensitive genes implicated in craniofacial development could reside within that band. Yet our patient does not present the low-set, posteriorly rotated ears (Fig. 1). Her hand phenotype resembles the photograph reported by Lucusa and Fryns [2000] for the nail hypoplasia, yet she presents no syndactyly. She is mostly distinct for the absence of any joint, heart, urinary, and genital tract anomalies, that frequently make up the 10q26 pure distal monosomy syndrome [Lucusa et al., 2002; Scigliano et al., 2004]. She displayed, however, the striking behavioral traits of the 10q26-qter monosomic patients, that is, aggressive behavior and a suicidal attempt. Although this episode followed a traumatic familial event, the father reported her to have frequent mood tantrums and her attention span was limited.
| 10q26-qter dela | 10q26-qter dupb | 10q25.1–q26.3 inv dup/10q26.3-qter delc | Present case 10q26.11–q26.2dup/10q26.22–q26.3 del | |
|---|---|---|---|---|
| Perinatal features | ||||
| Low birth weight | + | − | + | + |
| Microcephaly | + | − | − | − |
| Respiratory distress | + | − | − | + |
| Hypotonia | + | − | − | + |
| Strabismus | + | − | − | + |
| Facial dysmorphisms | ||||
| Long, triangular face | + | − | − | + |
| Facial asymmetry | + | − | − | + |
| Hypertelorism/epicanthus | + | − | − | + |
| Blepharophimosis | − | + | + | − |
| Prominent/broad nasal bridge | + | − | − | + |
| Long philtrum | − | + | − | − |
| Malformed/low-set/posteriorly rotated ears | + | + | + | − |
| Micrognathia | − | + | − | − |
| Other dysmorphic features | ||||
| Short neck | + | + | + | + |
| Joint contractures/camptodactyly | + | + | − | + |
| Lordosis/scoliosis | + | + | + | − |
| Hernias | + | − | − | − |
| Congenital heart defect | + | + | − | − |
| Urinary tract anomalies | + | + | − | − |
| Genital anomalies | + | − | − | − |
| Hands/feet defects | + | + | + | + |
| Follow-up | ||||
| Hypermobility | − | + | + | − |
| Convulsive seizures | + | − | − | − |
| Behavioral anomalies | + | − | − | + |
| Outcome | ||||
| Psychomotor delay | + | + | + | + |
| Short stature | + | − | + | + |
| Conductive hearing loss | − | − | + | − |
There are very rare observations of pure 10q26-qter trisomy probably due to their association with a mild phenotype [Devriendt et al., 1999; Hou, 2003]. Our patient shares a small duplication fragment of the 10q26.11-q26.3 region. Yet, consistently with the trisomic patients, she presents a skeletal phenotype of short neck and bilateral sandal gap (Fig. 2E). A similar skeletal phenotype is also present in the patient reported by Carter et al. [2010] with a slightly larger duplicated segment. Thus, the current observation further supports the possible specific phenotypic effect of a FGFR2 whole-gene duplication (a gene, which encodes a receptor tyrosine kinase important for osteogenesis), located within band 10q26.13, as postulated by Carter et al. [2010]. Moreover, our patient and the one reported by Carter et al. [2010] share a pre- and post-natal growth delay, not present in the pure trisomic patients. We think this is an effect of the deletions comprised in the rearrangements of these two patients, although of different size. The distinct facial feature of blepharophimosis of the 10q26.2-qter patients is not present in our current case, so we propose that a blepharophimosis dosage-sensitive locus resides within the most telomeric band of 10q26.3, that is present in our patient [Devriendt et al., 1999].
In summary, we propose that the patient reported here presents a distinct phenotype that should be diagnosed differentially from the 10q26 pure distal monosomy and trisomy syndromes. In detail, this phenotype consists of the craniofacial dysmorphism and behavioral abnormalities typical of the monosomic patients along with the skeletal features commonly seen in 10q26 trisomies. Although the duplication size is greater than the one of the deletion, the deletion is associated with a more severe phenotypic effect, as shown by the phenotypic comparisons illustrated in Table III. The specific rearrangement can be suspected in a low birth weight neonate with the facial phenotype of 10q26.2-qter patients that is not microcephalic and shows brachytelephalangy and bilateral sandal gap deformity. Yet, even if suspected on clinical grounds, this diagnosis is ultimately possible only by array CGH methods. This patient's phenotype could also be due to variable expressivity, incomplete penetrance or epistatic interactions affecting a genomic condition that cannot, however, be proved, unless similar patients are reported.
Our case has a microdeletion undetectable by conventional cytogenetic techniques at the level of 450 bands. The SSR markers deleted in the proband cover an approximately 4 Mb genomic region at 10q26.2–q26.3 approximately 145 kb from the telomere (The UCSC Human Genome Project Working Draft, http://genome.ucsc.edu/). Thirty five protein coding genes (Supplementary Table S1, see supporting information online) and two non-coding RNAs are located in the deleted region.
The duplicated region 10q26.11–q26.2 contains 67 protein coding genes (Supplementary Table S2, see supporting information online) and 17 non-coding RNAs (including six micro RNAs), while the intermediate “no change” region (between the duplication and the deletion) contains only two protein coding genes.
An explanation of the clinical picture of our patient which is not typical of the del 10q26 reported syndrome could be the coexisting microduplication of region 10q26.11–10q26.2 or a ribosomal gene position effect.
The clinical symptoms of the 10qter deletion cases reported so far were due to larger deletions, which were visible cytogenetically [Lewandowski et al., 1978; Turleau et al., 1979; Mulcahy et al., 1982; Taysi et al., 1982; Evans-Jones et al., 1983; Zatterale et al., 1983; Shapiro et al., 1985; Curtis et al., 1986; Mehta et al., 1987; Vanlieferinghen et al., 1987; Fryns et al., 1989; Gorinati et al., 1989; Greenberg et al., 1989; Wulfsberg et al., 1989; Kogasaka et al., 1990; Schrander-Stumpel et al., 1991; Teyssier et al., 1992; Petit et al., 1998; Suzuki et al., 1998; Leonard et al., 1999; Mutoh et al., 1999; Tanabe et al., 1999; Waggoner et al., 1999; Lucusa and Fryns, 2000; Ogata et al., 2000; Lucusa et al., 2002; Wu et al., 2002; Scigliano et al., 2004].
Recently Goldsmith et al. [2010] reported a 3-year-old girl with global developmental delay, poor coordination, and ataxia, with no MRI abnormality of the posterior fossa in whom a high-resolution (105 K) CGH microarray identified a do novo deletion of the terminal 4.6 Mb of the long arm of chromosome 10, within cytogenetic band 10q26.3. This finding was confirmed by FISH analysis.
Possible mechanisms of the satellited non-acrocentric chromosomes
Mechanisms for the translocation of NORs and satellites to non-acrocentric euchromatic regions are currently not well known. Schmid et al. [1984] supports the theory of very similar or identical DNA sequences of the translocated chromosomal regions, which promote pairing and crossing over between these non-homologous regions. The same authors consider another possible theory: In the interphase nuclei, the heterochromatic regions of chromosomes 15, 22, and Y are arranged in very close spatial contact (“somatic pairing”), which makes a translocation between them more likely. In this way it might be explained why the satellite of a Yqs chromosome originates most often from chromosome 15 or 22.
More frequent are translocations of NORs to a terminal region of another chromosome resulting in a satellited non-acrocentric chromosome. In this scenario, the terminal 4q is the most frequently affected region of an autosome [Hawks-Arn et al., 1995; Miller et al., 1995; Guttenbach et al., 1999]. This could indicate a special preference of the terminal 4q to recombine with the short arms of acrocentric chromosomes. The “illigitimate” breakage and reunion that produce these rearrangements may be due to the position of chromosomal segments containing DNA sequences with a high degree of homology [Giacalone and Francke, 1992; Lyle et al., 1995]. However, case numbers are too small to support a specific 4q preference, and NORs also have been reported to be translocated to terminal regions of chromosomes 1 through 12, 15 through 18, 21, X, and Y (Table I) (for review see Hawks-Arn et al., 1995).
Two explanations may account for the low number of reported breaks within NORs. First, the reported numbers may reflect their rarity and second, they could be due to technical limitations resulting in difficulties to identify them correctly.
The manifestation of a ribosomal gene position effect may depend on the position of the translocation breakpoint in the acrocentric short arm. If the breakpoint occurs within the ribosomal genes, the effect might be different than if the breakpoint is outside the ribosomal gene cluster. Ectopic placement of ribosomal genes can result in both normal and abnormal phenotypes, as a result of a hypothetic variable influence on neighboring gene expression [Hawks-Arn et al., 1995].
On the basis of our observations and of several other reports, we could conclude that microdeletions and/or microduplications occur very often in non-acrocentric satellited chromosomes and micro-array CGH is the method of preference which reveals such abnormalities. It is possible also, on theoretical basis that a ribosomal gene position effect exists, but this is very difficult to prove.
As a result we encourage careful evaluation of families with rearrangements involving the translocation of ribosomal genes into euchromatic regions of the genome.
The publication of detailed clinical information about individuals with such imbalances, when they occur de novo, is quite important to delineate the clinical features associated with particular genes, and guide our counseling of parents in clinical practice.
We thus encourage the application of molecular cytogenetic methods (as FISH and array CGH) in the evaluation of satellited non-acrocentric chromosomes of new cases and the reevaluation of previously reported cases as these methods can reveal the existence of complex chromosome abnormalities as was proven in our patient.
Acknowledgements
The secretarial help of Ms. Anna Stamati and Mrs. Antigoni Souli as well as the computer assistance of Ms. Konstantina Merou and the technical advice concerning array CGH of Dr. Markos Mihalatos are gratefully acknowledged.




