A de novo supernumerary genomic discontinuous ring chromosome 21 in a child with mild intellectual disability†‡
Nicoletta Villa and Angela Bentivegna contributed equally to this work.
How to Cite this Article: Villa N, Bentivegna A, Ertel A, Redaelli S, Colombo C, Nacinovich R, Broggi F, Lissoni S, Bungaro S, Addya S, Fortina P, Dalprà L. 2011. A de novo supernumerary genomic discontinuous ring chromosome 21 in a child with mild intellectual disability. Am J Med Genet Part A 155:1425–1431.
Abstract
Small supernumerary marker chromosomes (sSMCs) are structurally abnormal extra chromosomes that cannot be unambiguously identified or characterized by conventional banding techniques alone, and they are generally equal in size or smaller than chromosome 20 of the same metaphase spread. Small supernumerary ring chromosomes (sSRCs), a smaller class of marker chromosomes, comprise about 10% of the cases. For various reasons these marker chromosomes have been the most difficult to characterize; although specific syndromes have not yet been defined, 60% of cases are associated with an abnormal phenotype. The chromosomal material involved, the degree and tissutal distribution of mosaicism, and the possible presence of uniparental disomy, are the important factors determining whether or not the ring chromosome will give rise to symptoms. Using conventional and molecular cytogenetics approaches we identified a de novo chromosome 21 sSRC in a child with speech delay and mild intellectual disability. By using aCGH analysis and SNP arrays, we report the presence of two discontinuous regions of chromosome 21 and the paternal origin of the sSRC. A thorough neuropsychiatric evaluation is also provided. Only few other cases of complex discontinuous ring chromosomes have been described in detail. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Small supernumerary marker chromosomes (sSMCs) are structurally abnormal extra chromosomes that cannot be unambiguously identified or characterized by conventional banding techniques and are generally equal in size or smaller than chromosome 20 on the same metaphase spread [Liehr et al., 2004]. According to the data reviewed by Liehr and Weise [2007], sSMCs were present in 0.075% of all analyzed prenatal cases but only in 0.044% of consecutively studied postnatal ones. In infertile subjects, 0.125% were sSMC carriers (male/female rate 7.5:1). In developmentally retarded patients the sSMC rate was elevated to 0.288% [Liehr and Weise, 2007]. The small size of sSMCs prohibits characterization by traditional banding techniques, and instead requires molecular cytogenetic techniques, such as FISH or array-based comparative genomic hybridization (aCGH), to determine the chromosomal origin.
sSMC are scientifically interesting due to an incomplete understanding of their formation, karyotypic evolution, and the fact that their presence may lead to chromatin imbalances (partial tri-, tetra-, or hexasomies) without detectable clinical consequences [Liehr et al., 2008]. Subjects with a supernumerary marker chromosome have duplication and, in some cases, a triplication of the material comprising the sSMC. The risk of an abnormal phenotype associated with a randomly ascertained de novo sSMC derived from acrocentric autosomes (excluding chromosome 15) is approximately 7%, compared with approximately 28% for sSMC derived from non-acrocentric autosomes [Crolla, 1998]. A collaborative study of 19 Italian laboratories involving 241 sSMCs [Dalprà et al., 2005] has allowed definitive conclusions concerning karyotype–phenotype correlations. These studies emphasize that molecular characterization of sSMCs and their associated clinical phenotypes can provide karyotype–phenotype correlations useful for genetic counseling.
We report on a de novo small supernumerary ring chromosome (sSRC) of paternal origin, derived from different regions of chromosome 21, in a child with speech delay and mild intellectual disability.
MATERIALS AND METHODS
Cytogenetics
Metaphase chromosome preparations were obtained from PHA-stimulated lymphocyte cultures of peripheral blood according to standard procedures. Chromosome analysis was carried out applying QFQ banding according to routine procedures, and karyotypes for the patient (50 metaphases) and for both parents (100 metaphases, each) were reconstructed following the guidelines of ISCN 2009 [Shaffer et al., 2009].
Fluorescence In Situ Hybridization (FISH)
FISH was performed as previously reported [Lissoni et al., 2009]. To characterize the sSMC, the following probes were applied: Pan-centromeric, Pan-telomeric, D13Z1/D21Z1, beta-satellite, and WCP for chromosome 21 (Oncor, Inc., Gaithersburg, MD). To define the chromosome 21-derived euchromatic sequences, BAC probes were applied: RP11-91N21, RP11-106K13, RP11-22D1, RP11-61A21, RP11-97K13, RP11-78J18, RP11-242C13, RP11-141K11, and RP11-89M24 (Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK).
Array-CGH
Genomic DNA was extracted from patients' lymphoblastoid cultures by phenol–chloroform standard protocols and DNA concentration was determined on a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Berlin, Germany). Gene copy number analysis was performed by aCGH using Agilent Human Genome CGH Microarray 44K kit (Agilent Technologies™, Walldbron, Germany) following manufacturer's recommendations. Genomic DNA (Promega™, Mannheim, Germany) was used as reference in sex-matched hybridizations which were analyzed using Feature Extraction v10.5 and DNA Analytics v4.0 software (Agilent Technologies™) applying ADM2 algorithm with a threshold of 5, minimum absolute average log 2 ratio in called intervals of 0.30 and a minimum of three probes. Putative chromosome copy number changes were defined by intervals of three or more adjacent probes and were considered as duplicated or deleted when results exceeded the ±0.30 range.
High-Resolution Cytogenetics Analysis
DNA samples were hybridized to the Affymetrix Cytogenetics whole-genome 2.7 M array and processed according to the manufacturer's recommended protocol (http://www.affymetrix.com). This platform interrogates approximately 2.7 million markers, providing high-density coverage across the entire genome. Cytogenetics data was processed using Chromosome Analysis Suite (ChAS) software from Affymetrix (Santa Clara, CA). ChAS software was used to process the raw data (.CEL files) and determine regions of copy number segment gains/losses as well as copy-neutral loss of heterozygosity (cn-LOH). Copy number segments were filtered to include only gains represented by at least 250 markers extending over 500 kb, and losses represented by at least 100 markers over a 200 kb region. Confidence levels were restricted to at least 85%. Following copy number analysis on the Affymetrix Cytogenetics platform, the source of the chromosomal duplication was determined from trisomic genotype calls in the chromosome 21 ring region, which were estimated using the Standardized Centered Allelic Ratio (SCAR). The SCAR value was computed from Affymetrix software, where positive values map to the homozygous “A” allele, negative values to the homozygous “B” allele, and values close to 0 indicate heterozygosity. In duplicated regions, positive SCAR values indicate duplication of the “A” allele and negative SCAR values indicate duplication of the “B” allele. Heterozygous alleles in the proband where one parent carried the homozygous “A” allele and the other parent carried the homozygous “B” allele provided evidence to determine the source of the extra copy.
Sequence In Silico Analysis
In silico sequence analysis was performed using the following database and bioinformatic tools: Entrez Nucleotides Database, http://www.ncbi.nlm.nih.gov/Entrez/query.fcgi?db=nucleotide; NCBI BLAST, http://www.ncbi.nlm.nih.gov/Blast/; UCSC Genome Bioinformatics, http://www.genome.ucsc.edu/ (for sequence homology analyses for identification of gene content and for BAC/PAC clones used as probes in the FISH experiments).
CLINICAL REPORT
The female patient was born at 36 weeks of gestation after an uneventful pregnancy, as the second child of healthy, unrelated parents. The mother and father were 17 and 25 years old, respectively, and had a healthy older female child. Birth weight of the patient was 2,370 g with an Apgar score of 7 at 1 min. She was cytogenetically studied because of mild facial dysmorphisms and hypotonia. The family pedigree shows a history of Down syndrome (DS) (Supplemental Fig. 1). At 2 months, she was hospitalized for a suspected non-febrile seizure; weight was 5,250 g (90th to 97th centile), length of 55 cm (25th to 50th centile). Clinical evaluation confirmed a general muscular hypotonia, malformed ears, umbilical hernia, and gastroesophageal reflux, but neuropsychiatric evaluation, EEG, and MRI of the brain were normal. At 5 months her weight was 11,700 g (>97th centile). She was later referred because of slight delay in psychomotor development, especially speech delay. She could sit without support at the age of 7 months, and walk without support at the age of 20 months.
Neuropsychiatric Evaluation
The patient was referred for clinical assessment at the local Child Neuropsychiatry Service at the age of 29 months. The first psychomotor assessment (Brunet Lezine Test) showed a disharmonic psychomotor retardation: 20.8 months of developmental quotient (DQ) compared to 29 months of chronological age, with posture control (DQ 24 months), coordination (DQ 24 months), socialization (DQ 21 months), and significant impairments in the language domain (DQ 10 months). At the time of the psychomotor development check up at 39 months of chronological age the disharmonic psychomotor retardation was confirmed (DQ 24.9 months), with small improvement in all developmental domains, but with an increased retardation compared to the standardized average development. The differences between chronological and psychomotor age shifted from 8.4 months at the first psychomotor assessment (29 months of chronological age) to 14.3 months at the second check up (39 months of chronological age).
The patient underwent psychomotor therapy from age 4 to 6 years and speech therapy from age 6 to 8 years. When the speech therapy began, she showed difficulties with attention, language comprehension, and critical reasoning, and she was able to read by syllabicating. At the end of the speech therapy, at the age of 8, she was able to perform syllabic fusion, though decoding was very slow. The language, which had shown expressive delay, was simply structured and phonologically adequate, but lexical and semantic limits were more evident. Comprehension was limited to concrete contexts or related to personal experiences.
Results of the Leiter-R and speech assessment confirmed a mild intellectual disability with difficulties with logical, communicative and language abilities, attention, spatial memory, and short- and long-term memory (see a full report on Supplemental Material: Complete Neuropsychiatric Evaluation).
METHODS AND RESULTS
Chromosome analyses from the patient's blood lymphocyte cultures showed a sSMC, which was DA/DAPI negative, in all 50 metaphases investigated (Fig. 1a), while analysis of both parents showed normal karyotypes (100 metaphases investigated). FISH analysis using the Pan-centromeric probe and the Pan-telomeric probe (Oncor, Inc.) revealed a small monocentric sSRC, positive for the alphoid probe D13Z1/D21Z1, but negative for the beta-satellite probe (Fig. 1b–d). Finally, the WCP probe for chromosome 21 was positive demonstrating chromosome 21 origin and suggesting additional euchromatic sequences in the r(21) (Fig. 1e). Using ISCN 2009 recommendation, the aberration was characterized as: 47,XX,+mar de novo.ish r(21) (wcp21+,D13Z1/D21Z1+,βsat-).
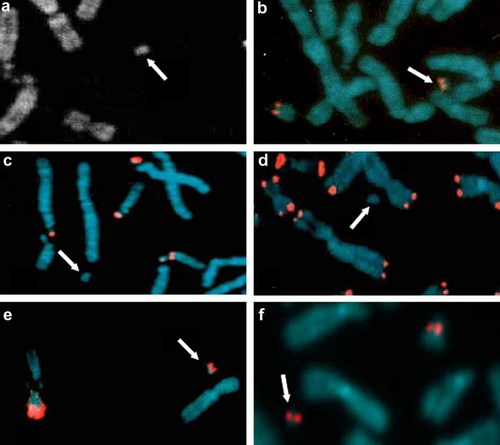
Results of cytogenetic and molecular-cytogenetic investigations showing a de novo supernumerary ring chromosome 21 in the proband. a: QFQ banding; (b) alphoid probe D13Z1/D21Z1; (c) beta-satellite probe; (d) Pan-telomeric probe; (e) WCP probe for chromosome 21; (f) RP11-106K13 BAC clone. The white arrows indicate the r(21) while the red signals indicate presence of the various probes.
In order to define the chromosome 21-derived euchromatic sequences and attempt to identify the breakpoint region, FISH analysis with a battery of BAC clones was performed on EBV-transformed lymphoblastoid cell line of the patient (Fig. 1f and Supplemental Fig. 2). The r(21) showed hybridization signals for RP11-97K13 and RP11-78J18, but none for RP11-141K11, while for RP11-242C13 there is no clear interpretation, since it showed a weak hybridization signal or no signals. From these results we were able to define a partial trisomy of about 3 Mb extending from cen to 21q21.1, with a breakpoint region of 158 kb by the size of RP11-242C13 (from nt 15,546,175 to 15,704,232). By aCGH analysis, two different regions of copy gain on chromosome 21 were evidenced (all the positions on chromosome 21 were referred to the old Assembly Mar. 2006 hg18 of UCSC): (a) the proximal one's, from 21q11.2 to 21q21.1 (nt 13,548,935–15,510,501), extending for 1.96 Mb; (b) the distal one's, from 21q21.3 to 21q22.11 (nt 29,278,776–31,123,552), extending for 1.84 Mb (Fig. 2). The two regions of copy gain were confirmed during subsequent whole genome analysis on the Affymetrix Cytogenetics array (Supplemental Fig. 3). Copy number segment analysis using the higher resolution cytogenetics array identified the amplified regions as 21q11.2–21q21.1 (1,066 markers; nt 13,527,258–15,552,772) and 21q21.3–21q22.11 (2,613 markers; nt 29,265,617–31,950,671).
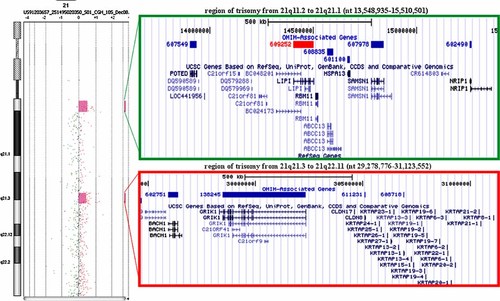
Array-CGH profile of chromosome 21 showing the two regions of trisomy present in r(21): the centromeric region covers 1.96 Mb, encompassing from 21q11.2 to 21q21.1 (nt 13,548,935–15,510,501) while the distal region covers 1.84 Mb, encompassing from 21q21.3 to 21q22.11 (nt 29,278,776–31,123,552). The colored boxes show magnification of the two regions with their known gene content.
Uniparental disomy (UPD) was excluded using microsatellite analysis (data not shown). These results were confirmed by the Affymetrix Cytogenetics array, which determined this region was free from long contiguous stretches of homozygosity (LCSH), which are characteristic of UPD. The paternal origin of the ring was assessed using allele signal intensities from the Affymetrix Cytogenetics Array. Trisomic genotypes were determined for the proband in regions 21q11.2 and 21q21.3–q22.11 and compared to the parental genotypes at locations where one parent carried the homozygous “A” allele and the other parent carried the homozygous “B” allele. The amplified allele in 24 out of 29 valid trisomic genotypes in this region matched the homozygous allele of the father (P < 0.001) (Supplementary Table I).
In Silico Sequence Analysis
In order to clarify the discontinuity of this ring chromosome, an in silico sequence analysis to delineate the genomic structural features of the two genomic regions involved was performed. The two regions of trisomy lie in the same contig (NT_011512: Homo sapiens chromosome 21 genomic contig, GRCh37.p2 reference primary assembly). BLAST analysis using the proximal region (as query) versus the distal one's (as subject), evidenced more than 30,000 blast hits on the query sequence, with a query coverage of 13% (see Supplemental Material: In silico Results, A). Furthermore, variation and repeats analysis by UCSC Genome Browser on Human Mar. 2006 (NCBI36/hg18) assembly was conducted for both regions. In the proximal region (chr21:13,548,935–15,510,501) several duplications of >1,000 bases of non-repeatmasked sequence were evidenced; three of them (chr21:28204081, chr21:28321445, and chr21:28335703) have a genomic size of 94,550 bases, 9,791 and 20,456 bases, respectively, and a similarity >96% with sequences belonging to the distal region of trisomy (chr21:29,278,776–31,123,552) (see Supplemental Material: In silico Results, B). Finally, again for the proximal region, The database of chromosomal imbalance and phenotype in humans using ensembl resources (DECIPHER) ([email protected]) reported one patient (249224) with a deletion of 6.37 Mb (chr21:14320039–20690911) affected by speech delay.
DISCUSSION
Here we report on the first case of a supernumerary discontinuous ring chromosome 21. However, this is the first instance where the sSRC was characterized in detail by FISH, array-CGH and then confirmed by the Affymetrix Cytogenetics array, as a de novo ring from chromosome 21. Previously, one case of ring chromosome originating from three discontinuous regions of chromosome 4 [Fang et al., 1995], and a second from different parts of the q arm of chromosome 20 [Anderlid et al., 2001] were described. Both reported the ring chromosomes were formed during repeated breakage and reunion cycles as a result of the formation of interlocked rings during cell division [Fang et al., 1995; Anderlid et al., 2001]. Two similar cases of r(1)s were identified in the work of Callen et al. [1999], but the discontinuous regions of chromosome 1 were not well characterized. Röthlisberger et al. [2009], report a discontinous sSMC(18), while Starke et al. [2001] described an acquired one from chromosome 11. Similar cases can be found at http://www.med.uni-jena.de/fish/sSMC/00START.htm, like 08-W-p23.3/1-1 for chromosome 8 and 14-W-q13.3/1-1 for chromosome 14. Although several discontinuous sSMCs have been reported, few were the sSRCs accurately studied.
In our case, we evidenced two different regions of partial trisomy on the q arm of chromosome 21 by array-CGH and also confirmed by the Affymetrix Cytogenetics array. Starting from Affymetrix Cytogenetics array data and microsatellite analysis it is possible to postulate a mechanism of formation for r(21) as initiating from a complete trisomy 21, originated more likely through a non-disjunction event at the second meiotic paternal division, or, less likely, at an early post-zygotic division. Then, in early stages of mitotic division, the supernumerary chromosome was reduced to a ring. The loss of genomic material from r(21) occurred by the mechanism proposed by Fang et al. [1995], or through non-allelic homologous recombination (NAHR), Indeed, NAHR can be mediated by the presence of low-copy repeats (LCRs, also called segmental duplications, SD) [Stankiewicz and Lupski, 2002; Shaw and Lupski, 2004]. LCRs are region-specific DNA blocks usually of 10–300 kb in size and of >95–97% similarity to each other [Stankiewicz and Lupski, 2002; Bailey and Eichler, 2006]. We evidenced the presence of at least three SDs in the two regions of trisomy, that may have triggered the mechanism NAHR, functioning as substrates (see Fig. 3).
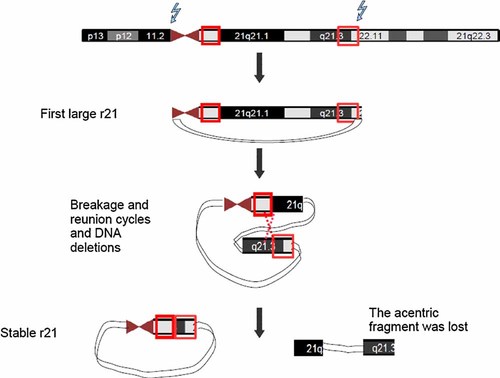
Proposed mechanism for the formation of the de novo supernumerary discontinuous r(21) from a larger ring. The two regions of trisomy of r(21) evidenced by array-CGH are highlighted by red rectangles. The red dashed lines represent a likely NAHR event between the two sub-regions with sequence homology nearly 100% identified by in silico analysis. See also BLAST analysis in Supplemental Material. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Finally, as the paternal lineage shows two relatives with DS, one could assume that there is a tendency to non-disjunction of the two chromosome 21s at meiosis in this family.
It is the distal trisomy region of r(21), not identified by FISH analysis and with higher gene content, to fall in the critical area associated with severe clinical problems (nt 18,100,000-q-tel) [http://www.med.uni-jena.de/fish/sSMC/21.htm#Start21]; this is in accordance with the model proposed by Liehr, based on the interpretation of the presently available data on sSMC and on the hypothesis that a genetic imbalance induced by sSMC presence, is the major reason for clinical symptoms in sSMC carriers. Supplementary Table II and Supplemental Figure 6 show an attempt of genotype–phenotype correlation in sSMC cases derived from chromosome 21 reported in the literature, in addition to the one presented here.
Our case emphasizes the difficulty of a genotype–phenotype correlation for cases with partial trisomy 21, as already discussed by others [Kondo et al., 2006; Lyle et al., 2009]. Kondo and coworkers reported two cases of partial trisomy 21 (pter-q22.12 and pter-q22.11) without the major features of DS. Both patients had a similarly sized extra chromosome 21, lacked all of DS critical region (DSCR) on 21q22, but they presented with moderate intellectual disability, delayed motor development, and speech delay, as our patient. Although the two cases described by Kondo et al. was associated with a distal partial trisomy 3p and 14q, respectively, the authors conclude that the moderate intellectual disability most likely results from partial trisomy 21pter-q22.1. On the other hand, Lyle and coworkers reported on genotype–phenotype correlations in DS in 30 cases of partial trisomy 21. Patients with milder phenotype are those in which the partial trisomy overlaps with one of the two regions of trisomy identified in our ring. From all this evidence one might speculate that a critical region, associated with psychomotor and intellectual disability and speech delay, maps to 21q22.11.
In conclusion, we were able, using aCGH analysis and SNP array, to identify the real genomic content of the r(21) in our patient, to determine its paternal origin (with important implications for counseling), to correlate with the phenotype and to hypothesize a possible mechanism of formation. It is increasingly evident that, given the rarity of these partial trisomies of chromosome 21, there is a need for a web-based collection of cases with both phenotypic and genotypic characterization, as suggested by Lyle et al.; in addition the use of oligonucleotide platforms for array-CGH may reveal not only additional cases that could be undiagnosed or better map the breakpoints, but also may help to define which regions contribute to phenotypic features.
Acknowledgements
We are grateful to the family for agreeing to participate in this study. We wish to thank the Galliera Genetic Bank for EBV-transformed lymphoblastoid cell line; Galbiati M. and Lettieri A. (M. Tettamanti Research Center, Pediatric Clinic, University of Milano, Bicocca, San Gerardo Hospital), Colombo D. (Istituto Auxologico Italiano) and Surrey S. (Thomas Jefferson University, Philadelphia, PA, USA) for technical support and editorial assistance. This work was supported in part by Fondazione Cariplo, AIRC, and MIUR (S.B., M. Tettamanti Research Center).




