A partial trisomy 1q patient with a deletion 1q22 and an insertion 1(q42q44) into 1q22†
How to cite this article: Misceo D, Rocchi M, van der Hagen CB, Frengen E. 2009. A partial trisomy 1q patient with a deletion 1q22 and an insertion 1(q42q44) into 1q22. Am J Med Genet Part A 149A:290–293.
To the Editor:
We report on a patient whose karyotype was defined to be 46,XY,der(1).ish del (q22) ins (q22q44q42) dup (q42q44), by combining G banding, aCGH, and FISH studies. As a child the patient had distinctive features, with a broad flattened nasal bridge, slight hypertelorism, downward slanted eyes, bushy eyebrows, divergent strabismus, trigonocephaly with a prominent forehead, short stubby nose, triangular mouth, high palate, long prominent philtrum and hypognathia (Fig. 1a). Birth weight is not known. He started to walk late and speech was delayed and difficult to understand. As a grown-up he has a long narrow face, is friendly and pleasant with a slurred pronunciation, functioning at about IQ 50–60. He has low hemoglobin with high transferrin values, but no known heart defects. Otherwise he is healthy with normal stature. He is now 53 years old, he lives in an institution and he is sociable.

a: Facial features of the propositus at the age of 2 years. b: Detail of the patient's karyotype (G-banding) showing chromosome 1 and der(1).
G-banding detected an abnormal chromosome 1 (Fig. 1b), which apparently contained a duplicated region in 1q24. aCGH (44K oligo array, Agilent Technologies, Santa Clara, CA; data analysis by BlueFuse, BlueGnome, Cambridge, UK) detected three aberrations: (1) 1p36.13 deletion (chr1:16686959–16794038 bp), (2) 1q22 deletion (chr1:152396245–152647439 bp), (3) 1q42–q44 duplication (chr1:221209415–245422360 bp) (supporting information Table I may be found in the online version of this article). The positions of the aberrations refer to the NCBI Build 35. The deletion in 1p36.13 is a well known copy number variation, detected in several independent studies (http://projects.tcag.variation), and it does not overlap with the region previously reported to be involved in the 1p36 microdeletion syndrome [Heilstedt et al., 2003]. This deletion was not verified by FISH because we assume that it does not contribute to the pathological phenotype in our patient.
The 250 kb deletion in chromosome 1q22 was confirmed by FISH using the BAC clones RP11-586I23 and RP11-243J18 which gave signal on chromosome 1, but not on der(1) (Fig. 2a,b and Table I). This region includes six genes: one gene encoding a hypothetical protein in addition to the genes; ASH1L, MSTO1, YY1AP1, DAP3, and GON4L. Because of the limited information about the function of these genes, we are not able to hypothesize the contribution of this deletion to the clinical phenotype of the patient.
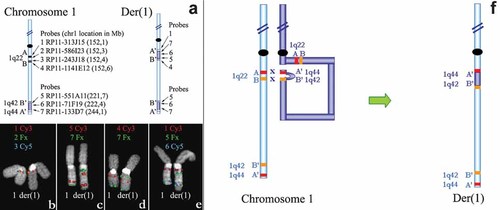
a–f: FISH analysis of der(1). a: The position of the probes used are indicated on the schematic drawing of chromosome 1 and der(1). b: Probe 1 (red, Cy3), Probe 2 (green, Fx), and Probe 3 (blue, Cy5) (See subpart a for probe names). Blue and green probes are deleted on der(1), while the clone in red flanks the deletion proximally. c: Probe 5 (red, Cy3) and Probe 7 (green, Fx). The orientation of a segment containing these probes is inverted in the centromeric location on der(1). d: Probe 4 (red, Cy3) and Probe 7 (green, Fx). The red probe flanks distally both the deletion and the duplication in the centromeric location on der(1). e: Probe 1 (red, Cy3), Probe 5 (green, Fx), and Probe 6 (blue, Cy5). The red probe flanks the duplication proximally. f: A hypothesis for the crossing over events resulting in the der(1) in the patient (details in the text).
| FISH probe (BAC) | Chromosome | Start (bp) | End (bp) | FISH signal |
|---|---|---|---|---|
| RP11-313J15 | 1q22 | 152147902 | 152354572 | Normal |
| RP11-586I23 | 1q22 | 152314231 | 152500224 | Deleted |
| RP11-243J18 | 1q22 | 152440697 | 152598904 | Deleted |
| RP11-1141E12 | 1q22 | 152598924 | 152777982 | Normal |
| RP11-1N12 | 1q42.12 | 221555884 | 221712258 | Normal |
| RP11-55A11 | 1q42.12 | 221702669 | 221847418 | Duplicated |
| RP11-71F19 | 1q42.12 | 222432705 | 222601611 | Duplicated |
| RP11-133D7 | 1q44 | 244095881 | 244262963 | Duplicated |
| RP11-103M14 | 1q44 | 244440460 | 244584199 | Duplicated |
| RP11-46P16 | 1q44 | 244728245 | 244900833 | Normal |
The third aberration detected by aCGH is a 23 Mb duplication. Even though the G-banding result indicated a duplication in chromosome 1q24, the duplication detected by aCGH maps in chromosome 1q42.1 → q44. FISH experiments revealed that (1) the duplicated segment (from RP11-55A11 to RP11-103M14, see Table I) is inserted in 1q in a proximal region and (2) it has inverted orientation compared to its original location (Fig. 2c). We then hypothesized that the duplicated 1q42.1 → q44-segment is inserted in 1q22, in connection with the deletion. The FISH results confirmed this hypothesis, since the two clones flanking the deletion (RP11-313J15 and RP11-1141E12) are located at the borders of the duplicated region in this patient (Fig. 2d,e).
No close relatives are available for investigations, and there is no family history suggesting the presence of inherited chromosome abnormalities.
The 1q22 deletion has a nearly perfect overlap with the SD1q22 region [Kuryshev et al., 2006], consisting of two segmental duplications head-tail oriented, spanning about 240 kb originated from an Alu-Alu mediated recombination about 37 million of years ago. The region contains approximately 40% Alu sequences, which is an enrichment compared to the genome average of 10.6% [Human genome consortium, 2001]. The abundance of Alu elements could facilitate the double crossover event resulting in the der(1) detected in our patient (Fig. 2f), which would involve two pairs of sequences: A and B delimiting the deletion in 1q22; A′ and B′ delimiting the duplicated region and located in 1q44 and 1q42.1, respectively. We used Blast2 (www.ncbi.nlm.nih.gov) to query for stretches of DNA, with high sequence identity and inverted orientation between A–A′ and B–B′, respectively. The best A–A′ candidates found were two 309 bp sequence elements with 96.4% nucleotide identity, with inverted orientation (supporting information Table II may be found in the online version of this article). These sequences corresponded to AluY type elements. The best B–B′ candidates found were two 307 bp AluYa5 sequences with 97% nucleotide identity (supporting information Table II may be found in the online version of this article). We suggest that the AluY members detected at the breakpoints are good candidates for the non-homologous crossing over leading to the aberrant chromosome 1 in our patient.
Several 1q partial trisomy patients have previously been described, but very few are considered pure 1q partial trisomies because of an involvement of a second chromosome. However, a distal 1q partial trisomy syndrome has previously been defined as patients with a proximal border of the trisomic region mapping telomeric to chromosome 1q41 [Duba et al., 1997]. Table II compares the clinical features of our patient to those reported for other cases of distal 1q trisomy [Chia et al., 1988; Bortotto et al., 1990; Verschuuren-Bemelmans et al., 1995; Concolino et al., 1998; Fan et al., 1999; De Brasi et al., 2001; Emberger et al., 2001; Cocce et al., 2007; Percesepe et al., 2007]. One common finding is the development delay/mental retardation affects 10/13 patients, however in only 2/13 this is reported as severe. Growth delay is often reported (9/13 patients), although this feature is not present in our patient. Minor anomalies, in particular prominent forehead (10/13 patients), long downward slanted eyes (8/10 patients) and prominent philtrum (6/13 patients) are also common manifestations. A more severe phenotype was described in three infants and one fetus, belonging to a large family, with a partial trisomy of 1q42–q44, due to an imbalanced segregation of a balanced translocation [Leisti and Aula, 1980]. The affected individuals showed a distinct dysmorphic appearance, pre- and post-natal growth delay, psychomotor delay, but in addition they had internal organ malformations and very short life span (maximum 10 months; spontaneous miscarriage at week 20). The discrepancy between these severe clinical scenarios and the milder cases described in later studies and in the current report might be due to the presence of additional cryptic imbalances or to an involvement of slightly larger chromosome bands in the patients with the most severe phenotypes.
| Karyotype | 46,XY,der(1) ins(1)(q22q44q42) del(1)(q42) dup(1)(q42q44), our patient |
46,XY,dirdup(1)(pter->q42.11::q42.11->qter) [Bortotto et al., 1990] |
46,XX,dirdup(1)(pter->q42.11::q42.11->qter) [Bortotto et al., 1990] |
46,XY,der(22)t(1;22) (q42;p11) [Chia et al., 1988] |
46,XX,dup(1)(q41q44).ish dup(1)(q42.2q43) [Cocce et al., 2007] |
46,XY,der(8)t(1;8) (q42.1;p23.3) [Concolino et al., 1998] |
46,XX,inv dup(1)(q44q42).ish (dup dup del 1)(q44q42) [De Brasi et al., 2001] |
46,XX, der(8)(8qter-8p23.3::1q41-qter) [Emberger et al., 2001] |
46,XX, der(5)(5qter-5p13.1::1q41-qter) [Emberger et al., 2001] |
46,XY,der(1)(qter->p36.13::q42.3->qter) [Fan et al., 1999] |
46,XY,der(9)t(1;9)(qter-q41::q34-pter) [Percesepe et al., 2007] |
46,XX,der(15)t(1;15) (q42;p11) [Verschuuren-Bemelmans et al., 1995] |
46,Xy,der(15)t(1;15)(q42;p11) [Verschuuren-Bemelmans et al., 1995] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin | U | IM | U | BMT | De novo | De novo | De novo | De novo | De novo | De novo | De novo | BPT | BPT |
| Broad flat nasal bridge | + | + | + | + | + | + | + | ||||||
| Short stubby nose | + | ||||||||||||
| Slight hypertelorism | + | + | + | ||||||||||
| Downward slanted eyes | + | + | + | + | + | + | + | + | |||||
| Bushy eyebrows | + | + | |||||||||||
| Divergent strabismus | + | ||||||||||||
| Prominent forehead | + | + | + | + | + | + | + | + | + | + | |||
| Triangular mouth | + | ||||||||||||
| High palate | + | + | + | ||||||||||
| Long prominent philtrum | + | + | + | + | + | + | |||||||
| Long narrow face | + | + | + | ||||||||||
| Slurry pronunciation | + | ||||||||||||
| DD/MR | IQ 50–60 | N | N | MiD | MoD | MiD | MiD | MiD | N | SeD | SeD | MiD | MiD |
| Growth | Normal | Delay | Delay | Delay | Delay | Delay | Delay | Delay | Delay | Delay | |||
| Age | 53 y | 3 y 1 m | Adult | 5.5 w | 4 y 5 m | 6 m | 2 y 3 m | 11 y | 2 m | 16 y | 12 m | 22 y | 20 y |
- U, known; IM, inherited from the mother; BMT, balanced maternal translocation; BPT, balanced paternal translocation; N, normal; MiD, mild delay; MoD, moderate delay; SeD, severe delay; y, year; m, month; w, week.
Acknowledgements
We are indebted to Eli Ormerod who performed the G-banding. This work was supported by project support from the University of Oslo. DM and EF were supported by Ullevål University Hospital Research Fund (VIRUUS).




