Molecular cytogenetic characterization of a 4p15.1-pter duplication and a 4q35.1-qter deletion in a recombinant of chromosome 4 pericentric inversion†
How to cite this article: Maurin M-L, Labrune P, Brisset S, Le Lorc'h M, Pineau D, Castel C, Romana S, Tachdjian G. 2009. Molecular cytogenetic characterization of a 4p15.1-pter duplication and a 4q35.1-qter deletion in a recombinant of chromosome 4 pericentric inversion. Am J Med Genet Part A 149A:226–231.
Abstract
To date, 10 cases of recombinant of chromosome 4 pericentric inversion involving sub-bands p14p15 and q35 have been described. We report on the first case analyzed using array-CGH in a female infant presenting psychomotor and growth retardation, facial anomalies, axial hypotonia, short neck, wide spaced nipples and cardiac defects. Conventional karyotype associated to FISH revealed a recombinant chromosome 4 with partial 4p duplication and 4q deletion derived from a paternal pericentric inversion. Array-CGH allowed us to precise rec4 breakpoints: the proposita carried a small 4.82–4.97 Mb 4q35.1 terminal deletion and a large 35.3–36.7 Mb 4p15.1 terminal duplication. Duplications of the distal 2/3 of short arm of chromosome 4 give rise to recognizable craniofacial features but no specific visceral malformation. A contrario small terminal 4q deletions are associated with cardiac defects. This case and review of literature suggest that two genes ArgBP2 and PDLIM3, located at 4q35.1 and both involved in cardiac and muscle development, could be responsible for cardiac defects observed in terminal 4q35.1 deletions. © 2009 Wiley-Liss, Inc.
INTRODUCTION
Gametogenesis in chromosomal pericentric inversion carriers may give rise to chromosomal recombination; recombination rate depending on the length of the inverted chromosomal segments. Genes contained in duplicated and deleted chromosomal segments determine morbidity and/or mortality affecting recombinant carriers. Duplicated 4p and deleted 4q recombinant of chromosome 4 pericentric inversion involving sub-bands p14p15 and q35 are viable [Gardner and Sutherland, 2004]. Up to now, nine cases of such chromosome 4 recombinant have been reported [Wilson et al., 1970; Dallapiccola et al., 1974; Rethore et al., 1974; Kleczkowska et al., 1992; Hirsch and Baldinger, 1993; Battaglia et al., 2002; Garcia-Heras and Martin, 2002; Stembalska et al., 2007].
We report on a 2-year-old female infant carrying a recombinant chromosome 4 of a paternal (p15.1q35.1) pericentric inversion, presenting with psychomotor delay, growth retardation, facial anomalies, short neck, increased inter-nipple distance, axial hypotonia, and cardiac defects. Breakpoints and size of duplicated and deleted segments of recombinant 4 were studied using array CGH.
CLINICAL REPORT
This female infant was the first child of healthy unrelated parents. She was born at full term, after an uneventful pregnancy. At birth, weight, height and head circumference were 3,290 g (50th centile), 45 cm (1st centile), and 35 cm (85th centile), respectively. Physical examination noted short neck, increased inter-nipple distance, mild edema of both feet, facial anomalies (anteverted nostrils, large philtrum, downslanting palpebral fissures, thin upper lip), and a systolic 3/6 murmur on heart auscultation. Mild axial and peripheral hypotonia was also noted. Turner syndrome was suspected and blood was sampled for chromosomes analysis. Echocardiography showed interventricular septal defect and interauricular septal defect. The infant was breast-fed. Both mother and daughter were discharged on day 7. At 2 months, weight and height were 4,430 g (15th centile) and 51cm (<1st centile), respectively; the infant was bottle-fed. Except for facial features, mild hypotonia, and systolic murmur, all previously noted, physical examination showed bilateral ovarian hernias, which were successfully operated on a few days later. At 13 months, weight was 9.5 kg (50th centile) and height was 68.5 cm (<1st centile); head circumference was 44.5 cm. The girl was able to sit by herself, to stand on all fours but was not able to walk. Axial hypotonia was obvious. Interventricular and auricular septal defects were unchanged. Ophthalmologic examination detected bilateral astigmatism and the girl was prescribed glasses. At 2 years, the child was able to walk alone. At 30 months, height was 81 cm (<1st centile), weight was 15 kg (85th centile) and head circumference was 48.5 cm (75th centile). Facial features were unchanged (Fig. 1). She was only able to say a few simple words. She was able to eat alone with a spoon. She was still hypotonic with bilateral convex pes valgus. Mild dorsolombar kyphosis was also present. Interventricular septal defect has spontaneously closed, whereas interauricular septal defect was unchanged. Educational support was continued.
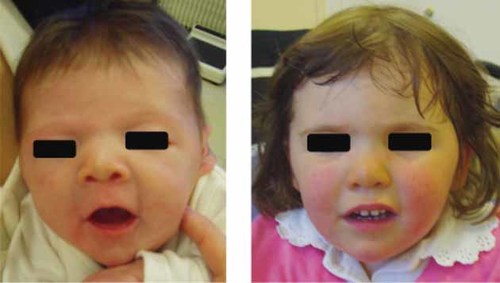
Proband at age 2 months (left picture) and 2 years (right picture). Note pug nose with anteverted nares, thin upper lip, and short neck. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
MATERIALS AND METHODS
Conventional Cytogenetic
Chromosomes analyses of proposita and her parents were carried out from cultured peripheral lymphocytes using RHG and GTG banding (Fig. 2).
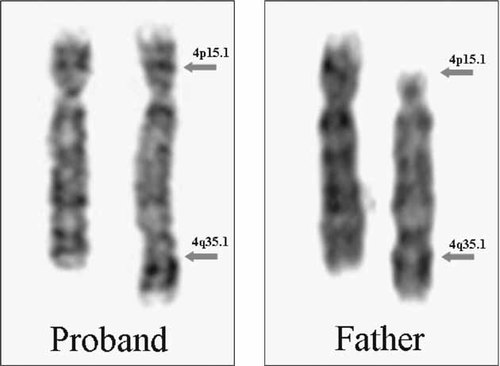
GTG banded chromosome 4 of proband and father. Grey arrows on recombinant chromosome 4 (proband) and inverted chromosome 4 (father) point to subbands 4p15.1 (short arm) and 4q35.1 (long arm).
Fluorescence In Situ Hybridization (FISH)
FISH analyses of proposita and father were performed on lymphocyte metaphase spreads. The following probes were used according to the manufacturers recommendations: chromosome 4 specific whole-chromosome paint (wcp) (QBIOgene, Illkirch, France), Wolf Hirschhorn syndrome critical region (WHSCR) (Abbott Molecular, Rungis, France), and 4p and 4q subtelomeric probes (AmpliTech, Compiegne, France). Specific 4p and 4q BAC probes were used to characterize the recombinant chromosome as listed in Table I.
| BAC clone | Location (bp) | Sub band | Proband FISH result | Father FISH result |
|---|---|---|---|---|
| RP11-1081C15 | 13,419–238,368 | 4p16.3 | dup | inv |
| RP11-399E10 | 3,042,767–3,243,146 | 4p16.3p16.2 | dup | np |
| RP11-326I19 | 4,832,556–5,036,138 | 4p16.2 | dup | np |
| RP11-623J3 | 25,595,749–25,760,824 | 4p15.2 | dup | np |
| RP11-20M7 | 32,138,110–32,292,383 | 4p15.1 | dup | inv |
| RP11-166H17 | 37,761,281–37,937,338 | 4p14 | nal | nal |
| RP11-380D23 | 111,660,447–111,862,073 | 4q25 | nal | nal |
| RP11-783P19 | 181,947,643–182,110,833 | 4q34.3 | nal | np |
| RP11-451F20 | 184,365,840–184,561,883 | 4q35.1 | nal | nal |
| RP11-279K24 | 187,170,026–187,293,316 | 4q35.1 | del | inv |
| RP11-33M11 | 187,700,615–187,901,338 | 4q35.2 | del | np |
| RP11-45F23 | 190,520,006–190,668,423 | 4q35.2 | del | inv |
- dup, two signals on the abnormal chromosome; del, no signal on the abnormal chromosome; nal, one signal normally located on the abnormal chromosome; inv, one inverted signal on the abnormal chromosome, np, not performed.
Array-CGH
DNA was extracted using QIAamp DNA Blood Mini Kit (QIAGEN, Courtaboeuf, France). Whole genome CGH array was performed with BlueGnome CytoChip v2 4400 BAC (565 kb median resolution) according to manufacturer's recommendations (BlueGnome, Cambridge, UK), scanned on Agilent DNA micro-array scanner (Agilent Technologies, Massy, France) and analyzed with BlueFuse software (BlueGnome, Cambridge, UK).
RESULTS
Conventional Cytogenetic
Conventional cytogenetic analyses revealed that the proposita carried an abnormal chromosome 4 with a q arm longer than usual.
Karyotype of the mother showed a normal female 46,XX formula. Karyotype of the father deomonstrated a chromosome 4 pericentric inversion 46,XY,inv(4)(p14q35). Secondary familial investigation revealed the father's inversion was inherited from his mother.
FISH
In situ hybridization using chromosome 4 specific whole chromosome paint showed that the additional material on the abnormal chromosome 4 of the patient was made of chromosome 4 sequences.
FISH using sub-telomeric 4q specific probes revealed that the recombinant chromosome 4 carried a sub-telomeric 4q deletion. FISH using subtelomeric 4p and WHSCR specific probes revealed a 4p duplication consisting in subtelomeric 4p probe and WHSCR probe hybridizing respectively on normal 4p loci and on the end of the long arm of recombinant chromosome 4.
BAC probes hybridization (Table I, Fig. 3) confirmed a terminal 4q deletion. The q arm breakpoint was located between non-deleted BAC RP11-451F20 (4q35.1) and deleted BAC RP11-279K24 (4q35.1), defining a 4–7 Mb terminal deletion. BAC probes hybridization also confirmed a 4p terminal duplication. The p arm breakpoint was located between non-duplicated BAC RP11-166H17 (4p14) and duplicated BAC RP11-20M7 (4p15.1), defining a 32–37 Mb terminal duplication.
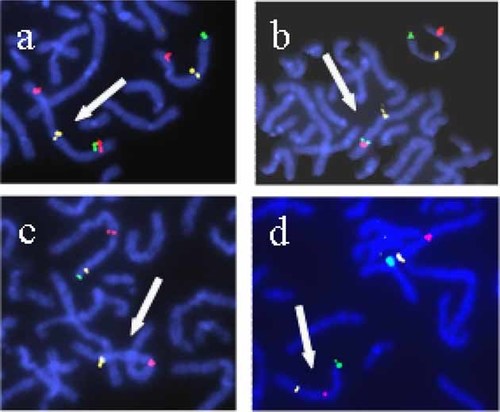
FISH studies of proband (a,c) and father (b,d). Same BAC probes were used for both proband and father: picture a and b show FISH performed with BAC probes RP11-451F20 (green), RP11-20M7 (red) and RP11-380D23 (yellow) and picture (c) and (d) show FISH performed with BAC probes RP11-279K24 (green), RP11-380D23 (red) and RP11-166H17 (yellow). White arrow: abnormal chromosome 4, recombinant (a,c), inverted (b,d). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
FISH on metaphase spreads from the father using same clones as for the proposita (Table I) confirmed his pericentric inversion involved the same regions (Fig. 3).
Array-CGH
Array-CGH (Fig. 4) showed gain of a 4p region from telomeric 4p clone RP11-46D11, located 0.15 Mb from telomere, to 4p15.1 RP11-135M12, located 35.3 Mb from telomere. Next clone on the array, 4p14 RP11-103J17, normally detected, was located 36.7 Mb from telomere.
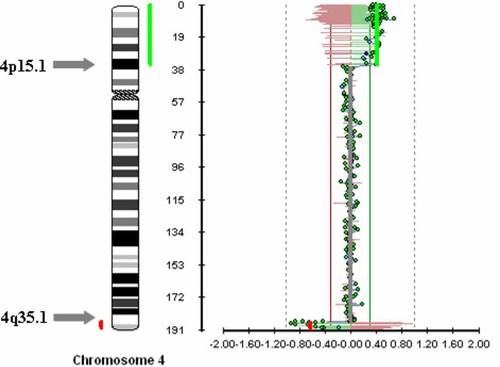
Array-CGH showing gain of a 4p15.1-4pter region and loss of a 4q35.1-4qter region. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Array-CGH also showed loss of a 4q region from telomeric 4q clone RP11-45F23, located 190.6 Mb from telomere, to 4q35.1 RP11-279O9, located 186.6 Mb from telomere. Next clone on the array, 4q35.1 RP11-28O17, normally detected, was located 186.45 Mb from telomere.
In summary, conventional and molecular cytogenetic investigations revealed that the proposita carried an unbalanced recombinant from a paternal chromosome 4 pericentric inversion resulting in a small, 4.82–4.97 Mb, 4q terminal deletion and a large, 35.3–36.7 Mb, 4p terminal duplication.
The patient's karyotype was 46,XX,rec(4)dup(4p)inv(4)(p15.1q35.1)pat.
DISCUSSION
We report on a 2-year-old female infant presenting with psychomotor delay, growth retardation, facial anomalies, short neck, increased inter-nipple distance, axial hypotonia, interventricular and interauricular septal defect associated with a recombinant chromosome 4 derived from paternal pericentric inversion. Partial 4p duplication and 4q deletion, suspected using conventional cytogenetic, were confirmed using FISH techniques. Breakpoints were localized using 0.5 Mb median resolution array-CGH. The proband carried a 46,XX,rec(4)dup(4p)inv(4)(p15.1q35.1)pat formula derived from her 46,XY,inv(4)(p15.1q35.1)mat father.
The first case of recombinant of a parental chromosome 4 pericentric inversion was reported by Wilson et al. 1970. We focused our literature review to recombinant cases of chromosome 4 inversion with breakpoints comparable to our case, that is, within sub-bands p14p15 and q35. Ten cases of such chromosome 4 pericentric inversion recombinant have been reported to date [Wilson et al., 1970; Dallapiccola et al., 1974; Rethore et al., 1974; Kleczkowska et al., 1992; Hirsch and Baldinger, 1993; Dufke et al., 2000; Battaglia et al., 2002; Garcia-Heras and Martin, 2002; Stembalska et al., 2007]. Clinical examination was not performed for one prenatal case [Dufke et al., 2000]. Two cases have been studied using FISH techniques [Stembalska et al., 2007] but none has been studied using array-CGH. Dufke et al. 2000 reported one family in which segregates a smaller pericentric inversion of chromosome 4. One pregnancy was terminated at 19 weeks of gestation because of unbalanced 46,XX,rec(4)inv(4)(p16.2q35.1)dup(4)(p16.2pter)pat karyotype detected after amniocentesis. Autopsy was not permitted. Father findings have been confirmed by FISH using WHSC specific probe but no molecular cytogenetic studies of the terminated pregnancy have been reported (data not included in Table II).
|
Wilson et al. 1970 |
Dallapiccola et al. 1974 |
Rethore et al. 1974 |
Kleczkowska et al. 1992 , patient 3 |
Hirsch and Baldinger 1993 , patient II-5 |
Battaglia et al. 2002 |
Garcia-Heras and Martin 2002 |
Stembalska et al. 2007 , older proband |
Stembalska et al. 2007 , younger proband |
Present case | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sex | m | f | m | f | f | m | f | f | f | f |
| Inversion | nr | p13q35 | p14q35 | p14q35.2 | p15.32q35 | p14q35 | p15q35 | p14q35 | p14q35 | p15.1q35 |
| Parental origin | pat | pat | pat | De novo | pat | mat | mat | mat | mat | pat |
| Prenatal growth retardation | + | + | + | nr | nr | + | nr | + | − | + |
| Postnatal growth retardation | + | + | + | nr | nr | + | + | − | − | + |
| Microcephaly | + | + | + | + | nr | + | + | + | + | − |
| Psychomotor retardation | + | + | + | + | + | + | + | + | + | + |
| Down slanted palpebral fissures | nal | + | nal | + | nr | − | − | nal | nal | + |
| Pug nose with anteverted nares | + | + | + | + | nr | + | + | + | + | + |
| Thin upper lip | + | + | + | + | nr | + | − | + | + | + |
| Pointed chin | + | + | + | + | nr | nr | − | − | + | |
| Abnormal ears | + | + | + | + | + | + | − | + | + | + |
| Short neck | nr | + | + | + | nr | + | + | + | + | + |
| Wide spaced nipples | nr | + | + | + | nr | nr | + | Broad chest | Broad chest | + |
| Dental anomalies | nr | nr | nr | + | nr | nr | Gingival hypertrophy caries | Delayed appearance of teeth caries | Delayed appearance of teeth caries | − |
| Coloboma | + | − | − | nr | nr | + | − | − | − | − |
| Congenital cardiopathy | − | − | + | nr | nr | + | + | − | − | + |
| Extremities abnormalities | + | +/− | + | + | nr | + | − | + | + | + |
| Genital abnormalities | + | − | + | nr | nr | + | − | − | − | − |
- +, present; −, absent; nal, normal; nr, not reported, m, male; f, female.
Comparing clinical features of the nine reported cases with the present one showed occurrence of: psychomotor retardation 10/10, microcephaly 8/10, anteverted nares 9/10, abnormal ears 9/10, thin upper lip 8/10, short neck 8/10, extremities abnormalities 8/10, prenatal growth retardation 6/10, postnatal growth retardation 6/10, wide spaced nipples 5/10, pointed chin 5/10, congenital cardiac defect 4/10, and genital abnormalities in males 3/3. No preferential parental origin 5pat/4mat/1 de novo was documented. Our patient showed psychomotor retardation, prenatal growth retardation, postnatal growth retardation, anteverted nares, abnormal ears, thin upper lip, short neck, extremities abnormalities, wide spaced nipples, pointed chin and congenital cardiac defect. Molecular cytogenetic studies allowed us to delimit rec4 syndrome critical region to terminal duplication of 4p15.1 and terminal deletion of 4q35.1.
Duplications of the distal 2/3 of short arm of chromosome 4 as reported here are associated with mental retardation (moderate to severe), prenatal and postnatal growth retardation, microcephaly, small forehead with heavy supraorbital ridges, prominent glabella, low hairline, deeply-set eyes, downslanted palpebral fissures, broad and flat nasal root with bulbous or pug nose tip, triangular mouth, high arched palate, large low-set posteriorly rotated ears, chin becoming elongated with age, short neck, widely spaced nipples, joint contractures, malposition of fingers and toes and genital anomalies for male patients [Kleczkowska et al., 1992; Schinzel, 2001; Battaglia et al., 2002]. Apart from nonspecific dysmorphic features, one major phenotypic difference between rec 4 syndrome patients and patients carrying a duplication of the same distal 2/3 of 4p appear to be the frequency of cardiac defects: 4/10 rec 4 patients suffer from congenital cardiac malformations, whereas cardiac defects are rarely observed in dup 4p patients [Patel et al., 1995; Schinzel, 2001].
Few pure terminal 4q deletions have been reported and 4qter terminal deletions are usually reported associated to various duplications [Tsai et al., 1999; Cingoz et al., 2006]. Cingoz et al. 2006 report 3 terminal 4q35 deletion cases including two relatives carrying same terminal del4q35.1/dup10p15. Common clinical features are: mental retardation (3/3), upturned nose (2/3), hypotelorism (2 (relatives)/3) malformed ears (3/3), asthma (2 (relatives)/3) and cardiac defects (atrial septal defect, ventricular septal defect, patent ductus, and pulmonary stenosis) (2/3).
Occurrence of cardiac defects in cases involving pure duplications of the same distal 2/3 of short arm of chromosome 4 such as the present case is rare [Patel et al., 1995; Schinzel, 2001]. It seems then reasonable to posit that the cardiac defects observed in rec4 patients are due to 4q deletion. As our patient also carried cardiac defects, we investigated in silico genes involved in cardiac development within her 4q35.1 terminal deletion. Two genes in the deleted region are involved in cardiac and muscle development: ArgBP2 and PDLIM3. ArgBP2 protein is involved in signal coordination and cell adhesion, motility, and structural organization of cardiomyocyte Z-discs [Taieb et al., 2008]. ArgBP2 is expressed 50- to 200-fold higher in heart than in other tissues and is detected on Z-disks of cardiac myofibrils of 7-day-old chicken embryo [Wang et al., 1997]. To our knowledge it has not been yet associated to cardiac malformations. PDLIM3, also known as ALP, is involved in upregulation during muscle differentiation and interacts and colocalizes with α-actinin at the Z-discs of striated muscle cells. ALP is expressed in cardiac muscle in mouse embryo and contributes to cardiac development and function. Disruption of ALP in mice gives rise to right ventricular dysmorphogenesis, trabeculation failure, and chamber dilatation [Pashmforoush et al., 2001; Pomiès et al., 2007].
We hypothesize that haploinsufficiency of ArgBP2 and/or ALP/PDLIM3 could disrupt embryonic cardiac development and be responsible for cardiac defects observed in terminal 4q35.1 deletions.
Acknowledgements
We are grateful to Laure Lecerf (IFR10-IM3, Créteil, France) for technical assistance.




