Functional disomy for Xq22-q23 in a girl with complex rearrangements of chromosomes 3 and X
Abstract
A 5-year-old girl with developmental and growth retardation is reported with complex chromosome rearrangements consisting of a partial Xq deletion and an abnormal chromosome 3 with multiple breakpoints. GTG-banding, and multiplex and conventional FISH studies showed that a 6.6-Mb Xq22-q23 segment was inserted into 3q, in addition to three intrachromosomal insertions in chromosome 3. Her karyotype was thus interpreted as 46,X,der(X)(Xpter→Xq22::Xq23→Xqter),der(3)(3pter→3p26::3p12→3q25.3::3p12→3p26::Xq22→Xq23::3q25.3→3qter). Replication R-banding study showed that the der(X) was inactivated in all blood lymphocytes analyzed. Methylation-specific PCR at the androgen receptor gene (HUMARA) locus at Xq11-q12 showed a skewed inactivation pattern with the active/inactive X chromosome ratio of 92/8. These data indicated the presence, in the majority of cells, of a functioning Xq22-q23 segment in both the normal X and the der(3) chromosomes. Her growth retardation, developmental delay, and other minor anomalies were most likely caused by dosage effects of the genes in the functionally disomic Xq22-q23 region. Despite the presence of two active copies of the proteolipid protein 1 gene (PLP1), she did not show the symptoms of Pelizaeus-Merzbacher disease, a subset of which has been known to be caused by the duplication of PLP1. © 2003 Wiley-Liss, Inc.
INTRODUCTION
Structural X-chromosome abnormalities may cause abnormal phenotypes in man. The X chromosome lacking XIST cannot undergo inactivation. A minute ring X chromosome sometimes escapes inactivation, resulting in functional disomy with some phenotypic effects [Jani et al., 1995; Migeon et al., 2000]. In females with a balanced X-autosome translocation, the derivative X chromosome containing a part of the autosome is usually active, and the remaining normal X chromosome is selectively inactive [Summitt et al., 1978]. On the other hand, if the derivative X chromosome is mainly inactivated, the severity of phenotypic abnormalities depends upon the number of somatic cells with functional disomy X in critical tissues [Mattei et al., 1982; Schmidt and Du, 1992], and upon extent of a disomic region of X chromosome and a monosomic region of autosome.
Here we report a girl with functional disomy for Xq22-q23 resulting from complex chromosomal rearrangements involving chromosomes 3 and X.
MATERIALS AND METHODS
Case Report
The patient, a girl, was born as the first child to a non-consanguineous, 26-year-old, G1P1 mother and a 30-year-old father, both healthy and phenotypically normal. Family history was unremarkable. Cesarean section was performed at 35 weeks' gestation because of oligohydroamnios, and fetal distress with tachycardia. Birth weight was 1,635 g (− 1.0 SD), length 40 cm (− 2.4 SD), and Apgar score 9 at 5 min. Features noted were a high palate, thick gingiva, abnormal dentition, low-set ears (Fig. 1a,b), nuchal hypertrichosis, bilateral short fifth fingers, left clubfoot, overlapping toes, hypervolemia, and hypotonia (Table I). Cardiac and abdominal ultrasonography and brain CT at age of 2 2/12 years showed no obvious abnormalities. Her developmental milestones were delayed: she lifted her head at the age of 9 months, sat alone at the age of 14 months, and walked without support at the age of 4½ years. At the age of 5 1/12 years, her height was 90 cm (− 4.0 SD), and weight 12.5 kg (− 2.2 SD). She spoke only several single-words. Her DQ was 27. MRI examination of the brain at the age of 5 1/12 years showed no evidence of dysmyelination. No clinical manifestations of Pelizaeus-Merzbacher disease were present, including microcephaly, jerking/rolling head movements, nystagmus, optic atrophy, ataxia, spasticity, or involuntary movements.
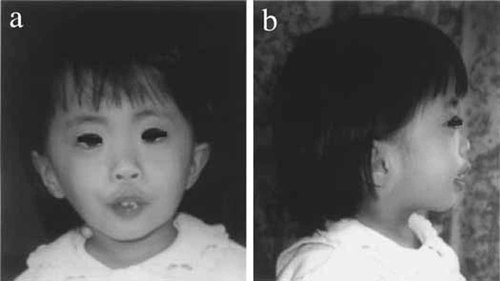
Frontal (a) and lateral (b) views of the patient at the age of 2 7/12 years.
| Clinical manifestations |
Carrozzo et al. [1997] |
Cremers et al. [1987] |
Schwartz et al. [1986] |
Steinbach et al. [1980] |
Present case |
|---|---|---|---|---|---|
| Sex | F | M | M | M | F |
| Karyotype | Dir dup | Dup | Inv dup | Dir dup | Ins(3;X) |
| Duplicated region | q21.32→q24 | q13→q22 | q21→q24 | q13→q22 | q22→q23 |
| X-inactivation | Random | Skewed | |||
| Duplication of PLP1 | + | NA | NA | +? | + |
| Oligohydroamnios | + | − | − | − | + |
| Fetal asphyxia | NA | − | NA | NA | + |
| IUGR | + | + | − | − | + |
| Neonatal jaundice | NA | + | NA | NA | − |
| Hypotonia | + | + | NA | + | + |
| Poor feeding | NA | NA | NA | + | − |
| Flat nasal bridge | NA | NA | + | + | + |
| High narrow palate | + | NA | + | NA | − |
| Micrognathia | NA | NA | + | NA | + |
| Down turned mouth | + | NA | + (carp shape) | − | |
| Nystagmus | + | + | NA | NA | − |
| Seizures | NA | − | NA | NA | − |
| Athetotic motor restlessness | NA | NA | NA | + | − |
| Growth retardation | + | + | + | + | + |
| Developmental delay | + | + | + (DQ < 50) | + | + |
| MRI/CT finding | Hypomyelination | Mild dilated ventricle, deficient myelination | NA | NA | Mild dilated ventricle |
| EEG abnormality | + | + | NA | NA | − |
| Abnormal funduscopy | + | NA | NA | NA | NA |
| Diagnosis of PMD | + (tentative) | + | NA | NA | − |
| Cardiac abnormality | NA | VSD, cardiac failure | NA | − | − |
| Hypoplastic genitalia | NA | NA | NA | + | − |
| Undescended testis | − | NA | + | + | − |
| Inguinal hernia | NA | + | NA | NA | − |
| Delayed bone age | NA | NA | NA | + | − |
| Clynodactyly of 5th finger | + | NA | NA | + | + |
| Overlapping toes | NA | NA | + | NA | + |
| Club foot | NA | NA | NA | NA | + |
| Scoliosis | NA | NA | NA | + | − |
| Skeletal abnormality | Hypoplastic pelvis | Dysplasia of pelvis | NA | NA | − |
| Agenesis of 12th rib | + | + | NA | NA | − |
- NA, not available.
Chromosome and FISH Analyses
G-banded chromosomal analysis was performed on cultured peripheral blood lymphocytes of the patient and her parents. Multiplex-FISH (M-FISH) analysis was adopted on the patient's chromosomes as described previously [Ida et al., 2002]. Conventional FISH studies using X chromosome-specific DNA probes were carried out in order to verify the results of M-FISH. Nine PAC/BAC clones mapped to Xq21.2-q26.3 (RP1-93L7, RP11-353J17, RP4-540A13, RP5-1070B1, RP11-81I3, PR1-300O13, RP3-404F18, RP3-417G15, and RP3-399M14) were obtained according to Map Viewer database (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/hum_srch?chr=hum_chr.inf&query). DNA was extracted from these clones using the plasmid midi kit (QIAGEN, Hilden, Germany), and labeled with SpectrumOrange™-dUTP or SpectrumGreen™-dUTP (Vysis, Downers Grove, IL) using the nick translation reagent kit (Vysis). The probes were hybridized to denatured chromosomal DNA of the patient at 37°C for 16 hr and washed. Photoimages of FISH were captured using a monochrome CCD camera (Carl Zeiss, Jena, Germany) on Axioplan 2 fluorescence microscope (Carl Zeiss) with appropriate filters and were analyzed with ISIS3 software (MetaSystems, Altlussheim, Germany).
X-Inactivation Study
Replication R-banding study was performed on chromosomes stained with DAPI as described elsewhere [Uehara et al., 2001]. FISH analysis was carried out using a PAC clone RP1-300O13 as a probe. Methylation-specific polymerase chain reaction (M-PCR) was carried out at the androgen receptor gene locus (HUMARA) at Xq11-q12, as described previously [Kubota et al., 1999]. In short, PCR products with primers specific for a methylated allele were compared quantitatively to those with nonmethylation-specific primers.
Gene Expression and Sequence Analyses
For gene expression analysis, two genes, PRPS1 (the phosphorybosyl pyrophosphate synthetase I gene) and DSIPI (the delta sleep inducing peptide gene), were used as representatives of expressed genes at the Xq22-q23 region in peripheral blood leukocytes. Two BAC clones, RP5-1070B1 corresponding to PRPS1, and RP11-81I3 corresponding to DSIPI, were used for FISH study to see whether they were present in inserted X chromosomal segment. To find informative polymorphisms, the parents' genomic DNA was sequenced for coding regions of the two genes, using an ABI 3100 automated sequencer (Perkin Elmer, Wellesley, MA).
RESULTS
G-banded chromosomes from the patient had complex chromosome rearrangements consisting of a partial Xq deletion and an abnormal chromosome 3 with multiple breakpoints (Fig. 2a). M-FISH revealed insertion of an Xq segment into the long arm of the abnormal chromosome 3 (Fig. 2b). FISH analysis using the nine X-chromosomal probes confirmed the insertion into der(3) and the deletion of der(X). Four clones (RP4-540A13, RP5-1070B1, RP11-81I3, and RP1-300O13) showed signals on the der(3), but no signal on the der(X) (Figs. 2c,d and 3). The other five clones (RP1-93L7, RP11-353J17, RP3-404F18, RP3-417G15, and RP3-399M14) showed signals on both the der(X) and normal X chromosomes (Figs. 2c and 3). The X-derived inserted segment was thus a 6.6-Mb region, 98.3-104.9 Mb proximal to Xpter according to the Map Viewer database. Re-examination of GTG-banded chromosomes indicated that der(3) had at least 3 breakpoints and 3p12→3q25.3, 3p12→3p26, and Xq22→q23 segments were inserted into bands 3p26, 3q12, and 3q25.3, respectively. Thus, the patient had a de novo 46,X,der(X)(Xpter→q22::q23→qter),der(3)(3pter→3p26::3p12→3q25.3::3p12→3p26::Xq22-q23::3q25.3→3qter) karyotype. The parents had normal chromosomes.
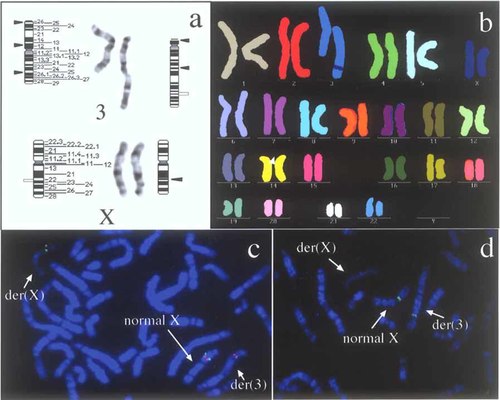
GTG-banding (a), multiplex-FISH (b), and conventional FISH using RP1-300O13 (Orange) and RP1-404F18 (Green) (c), and FISH using RP4-540A13 that corresponds to PLP1 on early-replicated R-banded chromosomes (d), in the patient. Arrowheads and out lined range indicate reunion points and inserted segment, respectively.
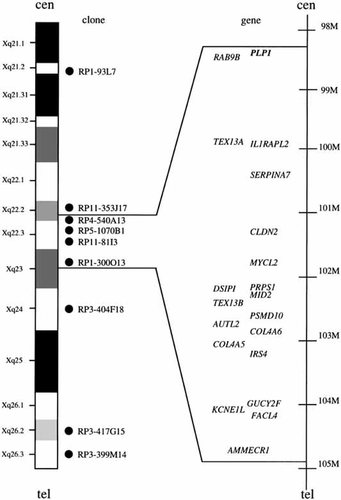
Schematic presentation of a 6.6-Mb inserted and deleted segment on der(3) and der(X) of the patient, respectively. Filled circles indicate clones used for FISH. Genes located to this segment are also indicated.
FISH analysis on R-banded chromosomes revealed that der(X) without any FISH signal for RP1-300O13 was always faintly stained and the normal X chromosome with a FISH signal was always well stained in all 100 cells analyzed (Fig. 2c), indicating that der(X) was consistently inactivated. M-PCR analysis at the HUMARA locus revealed a skewed inactivation pattern with the inactive/active X chromosome ratio of 92/8 (Fig. 4). There was no nucleotide change in the parents at the PRPS1 and DSIP loci. Therefore, further allelic expression studies were not carried out.
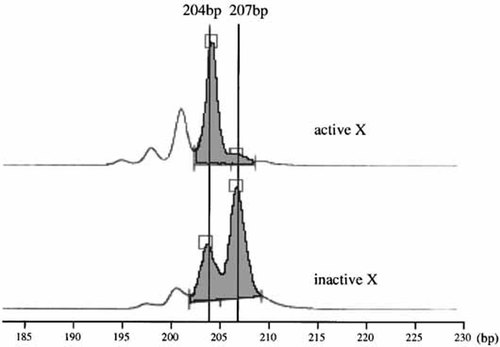
Methylation-specific PCR assay of HUMARA. Upper and lower rows show products of the patient using primers specific for unmethylated and methylated alleles, respectively.
DISCUSSION
Replication R-banding study and methylation-specific PCR analysis at the HUMARA locus of the der(X) in the patient revealed markedly skewed inactivation. The results in turn indicated that both the Xq22-q23 segment inserted into the der(3) and that present in the normal X chromosome were active, and is functionally disomic. The abnormal phenotype in the girl we described is thus likely to be attributable to a dose effect of the gene(s) in the Xq22-q23 disomic segment. Indeed, one or the other of hypotonia, growth and developmental delay, a high palate, down-turned mouth, delayed bone age, overlapping toes, and bilateral clinodactyly of 5th fingers was observed in any of four previously reported patients with a partial duplication involving the band Xq22 [Steinbach et al., 1980; Schwartz et al., 1986; Cremers et al., 1987; Carrozzo et al., 1997] (Table I). Disruption of gene(s) around the breakpoints in either or both chromosomes 3 and X also may have contributed to some of the manifestations in our patient.
Duplication of the proteolipid protein 1 gene (PLP1) merits comments. Of around a dozen genes known to be located at Xq22-q23 (the human genome browser, http://genome.ucsc.edu/cgi-bin/hgTracks) (Fig. 3), PLP1 (MIM 300401) is the only gene whose duplication could cause Pelizaeus-Merzbacher disease (PMD, OMIM No. 312080) and its allelic milder form, spastic paraplegia type 2 (SPG2). PMD is an X-linked dysmyelinating disorder of the brain with rotary movements of the head and eyes as one of first appearing symptoms, and spasticity of the limbs, cerebellar ataxia, and dementia developing later at around first and second decades of life. Duplications of PLP1 account for 50–75% [Hodes et al., 1993], and point mutations for 15–20% of PMD patients [Sistermans et al., 1998; Mimault et al., 1999]. Since functional disomy for Xq22-q23 observed in the girl we described includes the duplication of PLP1, symptoms of PMD may be expected. This is unlikely, because she never had any cardinal PMD symptoms. However, she is too young at the age of 5 years to be truly evaluated. As mentioned above, there have been four cases of a cytogenetically visible duplication for Xq22: a female with 46,X,dir dup(X)(pter→q24::q21.32→qter) [Carrozzo et al., 1997]; and three males each with 46,Y,dup(X)(pter→q22::q13→qter) [Cremers et al., 1987], 46,Y,inv dup(X)(pter→q24::q21→qter)mat [Schwartz et al., 1986], and 46,Y,dup(X)(pter→q22::q13→qter) [Steinbach et al., 1980] (Table I). Involvement of PLP1 with PMD features was confirmed in two cases at the age of 18 months and at 5 years, respectively [Cremers et al., 1987; Carrozzo et al., 1997], but not studied in the other two, though the male patient reported by Schwartz et al. [1986] should have had PLP1 duplication because of its large extent of the chromosomal duplication. However, no PMD features were present in the patient at the age of 16 months [Schwartz et al., 1986].
In conclusion, the girl we have described had a 6.6-Mb duplication at the Xq22-q23 segment that remains active. Despite the presence of two active copies of PLP1, she had no symptoms of PMD. Growth retardation, developmental delay, and other minor anomalies may be caused by a dosage effect of gene(s) in the functionally disomic X chromosomal region.




