Artificial intelligence generated leukemia cell images
Ngai Tung Eric Kwok and Stefan Winkler are senior authors who contributed equally to this work.
Artificial intelligence (AI) is a disruptive technology that holds great promise in medicine, due to its ability to effortlessly assimilate, integrate, and generate vast amounts of clinical data. In the hematology laboratory, the quality of digital blood cell images captured from peripheral blood films used in AI algorithms for blood cell classification1 far surpasses the early twentieth-century hand-drawn medical artwork of blood cells as seen under the light microscope (Image 1A). Advances in laboratory automation, coupled with high-throughput digital slide scanners allow the acquisition of high-resolution, whole-slide images of Romanowsky-stained peripheral blood films. This has enabled the creation of large image databanks of blood cells, allowing the exponential growth of artificial intelligence (AI) studies in blood cell classification and identification of blood-borne parasites such as malaria.2
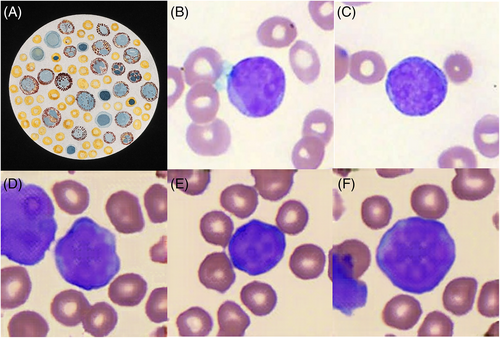
Departing from existing commercial laboratory solutions3 and conventional AI approaches in hematology focused on pattern recognition and task automation,4, 5 we describe the novel use of an AI generation algorithm to synthesize artificial, yet morphologically realistic images of leukemic blasts. We utilized an open-source AI StyleSwin algorithm6 which is based on a generative adversarial network (GAN), for the image synthesis of realistic leukemic blasts. A total of 21 739 pre-annotated digital images (for examples, see Image 1B, C) derived from peripheral blood films of our patients with acute myeloid leukemia (Wright stain, 80× objective, Motic EasyScan Infinity 60) were used in the training and evaluation process. A total of 17 431 images of leukemic blasts were used for training, with 4308 images randomly chosen for model evaluation. The annotation of each cell was performed by 2 laboratory-certified medical technologists and validated by a hematologist. The StyleSwin GAN model was trained with default parameters and a batch size of 3 on a Tesla V100 GPU with a graphics RAM size of 16 GB. Sample images were generated at every 50 000 steps for evaluation, and the model was trained with a total of 500 000 steps.
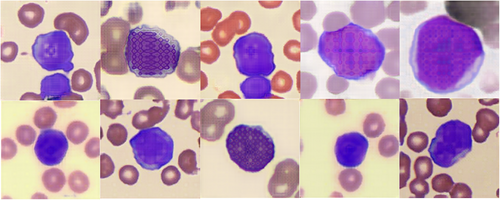
The AI-generated blasts had classic morphological features, which included a high nuclear-cytoplasmic ratio, the appearance of less condensed nuclei with an immature chromatin pattern, as well as the presence of prominent nucleoli7 (Image 1D–F). The AI-generated images of red blood cells in the image background were representative of the classic red cell appearance, with anuclear red blood cells with characteristic central pallor seen. The main limitation of the AI-generated leukemic blasts was that there was coarse pixelation of nuclei and nucleoli, whereas in digitized images of peripheral blood film-captured blasts (Image 1B, C), the fine chromatin pattern of the nuclei is seen without obvious pixelation. An explanation for the coarse pixelation may be due to either the image resolution limitations of StyleSwim's image generation method or the limited amount of GAN training performed; or both factors. Furthermore, other features that are characteristic of myeloid blasts such as Auer rods were not generated, even though the training dataset did have a small proportion of blasts with Auer rods (approximately 2% of all images). The AI model also generated a series of blasts that were less realistic (Image 2). These had irregular nuclei with a checkered pixelated pattern which were morphologically not consistent with myeloid blasts. However, the less realistic artificially created blasts still demonstrated some classic features, which include a high nuclear-cytoplasmic ratio, and in some cells, the presence of a crude, irregularly, pixelated nucleolus. This suggests that the GAN was able to assimilate and capture the key morphological features of blasts from the training dataset, but not able to generate highly accurate versions consistently.
AI-generated medical images using GAN8 can help overcome medical data scarcity and mitigate privacy concerns regarding the use of patient data. These images may potentially be enhanced with advanced computer algorithms or recreated with higher resolution with less noise and used to augment research datasets. Such images could also contribute to testing in computer-simulated benchtop trials for regulatory evaluation of AI software as a medical device (SaMD). Finally, AI-powered image generation offers a unique tool to facilitate the education of healthcare staff.9 In digital morphology, learners can be presented with images of one or more relevant cell types depicting existing morphological features to reinforce the structure appearance and key characteristics of different cells. Further frameworks are needed to evaluate the validity of AI-generated images for use in clinically meaningful endpoints and should be subjected to quality control by domain experts to reduce the risk of harm-by-data that result in algorithmic biases,10 before clinical application in patient care.
AUTHOR CONTRIBUTIONS
All authors contributed to the image curation, annotation, and analysis. Ngai Tung Eric Kwok performed the AI training and generation of digital images, with Stefan Winkler providing supervision and technical guidance.
FUNDING INFORMATION
Bingwen Eugene Fan is supported by the National Medical Research Council (NMRC) Clinician Innovator Development Award (NMRC/CIDA19May-0004). This work was funded in part by the Centre for Medical Technologies and Innovations—National Health Innovation Centre grant (CMTi-NHIC3-21-01-07).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




