Comprehensive analysis of TP53 mutation characteristics and identification of patients with inferior prognosis and enhanced immune escape in diffuse large B-cell lymphoma
Tingting Zhang, Yaxiao Lu and Xia Liu contributed equally to this work.
Funding information: Clinical Oncology Research Fund of CSCO, Grant/Award Numbers: Y-Roche20192-0097, Y-XD2019-162; National Human Genetic Resources Sharing Service Platform/Cancer Biobank of Tianjin Medical University Cancer Institute and Hospital, Grant/Award Number: 2005DKA21300; National Key New Drug Creation Special Programs, Grant/Award Number: 2018ZX09201015; National Natural Science Foundation of China, Grant/Award Number: 81770213; Natural Science Foundation of Tianjin City, Grant/Award Numbers: 18JCZDJC45100, 19JCYBJC26500; The Science and Technology Research Program of Tianjin Education Commission, Grant/Award Number: 2019KJ191
TP53 mutations have been observed in diffuse large B-cell lymphoma (DLBCL), with a mean frequency of ~20%. Studies on TP53 mutations as prognostic markers have historically been controversial, and the results have not been consistent across different studies on DLBCL. Considering the complex pathophysiological mechanisms involved in DLBCL, we wondered whether the interaction of TP53 with other genetic variants could further promote the development of DLBCL, and thus be more prognostically predictive. Moreover, whether the genetic interactions between TP53 and other oncogenic mutations could shape the discrepant immune landscape in DLBCL remains unknown, as these genetic alterations usually drive the malignant phenotype and directly or indirectly affect the tumor microenvironment (TME) and support tumor survival.
In this study, we performed a comprehensive analysis of the genomic characteristics of TP53 through high-throughput sequencing in patients with de novo DLBCL. Patients' characteristics are reported in Table S1. Detailed methods are provided in the Supplementary Material. A total of 227 significantly mutated genes were identified (Table S2), of which TP53 was the second most frequently mutated gene, with a rate of 30% (53 of 176) and 62 sequence variants detected. Among these variants, 74% (n = 46/62) were missense mutations, and the remaining were inactivating frameshift indels (n = 7), nonsense mutations (n = 3), coding sequencing indels (n = 4), and splicing mutations (n = 2). Mutation patterns and distributions are shown in Figure S1 and Table S3. Importantly, most mutations (56/64, 87.5%) occurred in exons 5–8, which encoded the DNA-binding domain (DBD) region of TP53 (Figure S1C,E). Codons 175, 273, and 248 of the p53 protein had the highest mutation frequency, which are also the hot spots of TP53 mutation found in most human cancers (Figure S1D). Given that the DBD of TP53 is the functional central core domain and mutations in this region potentially have a strong impact on TP53 function, we mainly focused on mutations in this region. Patients were divided into TP53-MUT and TP53-WT groups according to TP53 mutation status in the DBD region. There were no differences in the number of small deletions/insertions and single nucleotide variants (SNVs) between the TP53-MUT and TP53-WT groups (Figure S2). Moreover, the tumor mutation burden (TMB) was similar between the two groups (Figure S2E). Clinical relevance between TP53 mutation and clinicopathological characteristics, such as age, sex, B symptoms, stage, number of extranodal sites, performance status, LDH level, and International Prognostic Index was not observed (Table S4). Note that TP53 mutations significantly enriched in the GCB subtype (p = .033, Table S4). Among the 176 patients, 155 patients having the complete follow-up data were enrolled in the survival analysis. Overall, patients with TP53-MUT tended to have inferior overall survival compared with patients with TP53-WT (median: 92.3 versus 110.8 months, respectively, p = .17, Figure 1A), but it did not reach statistical significance. A subgroup analysis showed that the potential predictive value of TP53 mutations was mainly attributed to the GCB subtype (Figure S3).
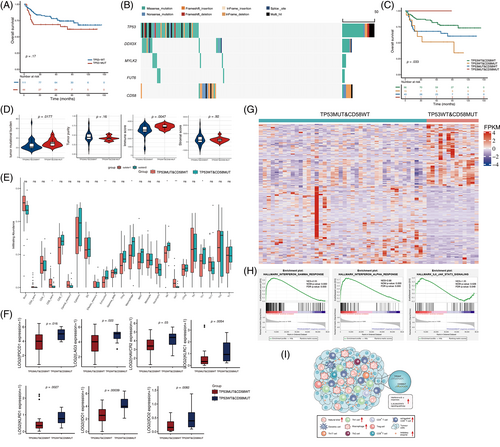
We next recognized the genomic variants that co-occur or are mutually exclusive with TP53. We observed that DDX3X, MYLK2, and FUT6 mutations co-occurred with TP53 mutations, and CD58 mutations were mutually exclusive with TP53 mutations (Figure 1B and Table S5). Specifically, patients were divided into four groups based on the mutation status of these genes. No significant difference was observed in survival among the three groups according to the combination of co-occurring mutation genes with TP53 (p = .37 for DDX3X; p = .11 for MYLK2; p = .54 for FUT6; Figure S4). However, we found that the combination of TP53 and CD58 mutations could significantly distinguish the prognosis of patients with DLBCL (p = .033, Figure 1C). Patients with both wild-type TP53 and CD58 had a better prognosis than patients with either of the two mutually exclusive modes of CD58 and TP53 mutations. Unexpectedly, patients with TP53 wild-type and CD58 mutations (TP53WT&CD58MUT) had worse survival than those with TP53 mutations and CD58 wild-type (TP53MUT&CD58WT). Because TP53 and CD58 mutations were mutually exclusive, only one patient harbored both TP53 and CD58 mutations, and the patient still alive at the last follow-up. The predictive value of TP53WT&CD58MUT group was also observed in patients with GCB-DLBCL (Figure S5). Moreover, the relationship of a mutually exclusive mutant between CD58 and TP53 and the prognostic significance of this interaction were validated using publicly available data from 1001 patients with DLBCL from the Duke University's cohort1 (Figure S6).
We then explored whether the cooperation of the mutually exclusive mutations between TP53 and CD58 may profoundly influence the microenvironment in DLBCL. We found that the overall TMB was significantly higher in the TP53WT&CD58MUT group than in the TP53MUT&CD58WT group (p = .0177, Figure 1D), while there was no difference in TMB when dividing patients only according to TP53 mutation status (p = .5348, Figure S2E). In addition, the ESTIMATE immune scores in the TP53WT&CD58MUT group were significantly higher than that in the TP53MUT&CD58WT groups (p = .0047) (Figure 1D). Moreover, the exhausted T cell, macrophage cell, NK cell, and Th1 cell enriched in the TP53WT&CD58MUT group (Figure 1E). The difference between the two groups was mainly due to the combined influence of the mutation pattern “TP53WT&CD58MUT,” but rather only affected by the CD58 mutations, given the immune cell infiltration was similar when dividing patients just according to CD58 mutation status (Figure S7). Furthermore, the co-inhibitory receptors such as PD-1, TIM3, and LAG3 were preferentially expressed in the TP53WT&CD58MUT group (Figure 1F and Table S6). However, there was no difference in the expression of co-stimulatory molecules (Table S6). Inhibitory immunomodulators were also significantly upregulated in the TP53WT&CD58MUT group when comparing with the TP53WT&CD58WT group (Figure S8), suggesting the unique immune phenotype in the TP53WT&CD58MUT group. The findings that high immune scores and abundant infiltrating exhausted T cells in the TP53WT&CD58MUT group were validated in an independent external cohort from the REMoDL-B trail (N = 400)2 (Figure S9 and Table S7).
Finally, we investigated the differentially biological pathways between the TP53WT&CD58MUT and the TP53MUT&CD58WT groups. Five hundred differentially expressed genes were identified with a false discovery rate less than 0.05 and |log2foldchange| > 1. One hundred genes were significantly upregulated and 400 genes were significantly downregulated in the TP53WT&CD58MUT group (Figure 1G). Figure S10 presented the enriched gene ontology terms in the TP53WT&CD58MUT group, including cytokine and chemokine production, binding and activity, and interferon-γ pathways. GSEA showed significantly activated interferon-α and interferon-γ responses and IL-6/JAK/STAT3 signaling in the TP53WT&CD58MUT group (Figure 1H). The immune landscape of patients with TP53WT&CD58MUT harbored was summarized and conceptualized in Figure 1I.
The value of TP53 gene alterations in predicting survival in DLBCL remains controversial, even in the era of sequencing. In the L.M. Staudt' study, four prominent genetic subtypes were identified based on the molecular classifications. Notably, TP53 was not significantly enriched in one of these subtypes, although TP53 was the much frequently mutated gene (25.2%). On the basis of this classification, L.M. Staudt and colleague further distinguished ST2, A53, and mixed subtypes, and the survival of A53, characterized by inactivation of TP53, was intermediate in this model.3 The C2 molecular subtype identified in Chapuy's study corresponding to A53 also showed a tendency for a poor prognosis.4 A subsequent study demonstrated that the impact of TP53 mutations on survival was relied on the genetic context of the lymphoma, conferring no effect in the SOCS1/SGK1 clusters and NOTCH2 subtype and inferior prognosis in the MYD88 subtype.5 These results demonstrated that TP53 alterations have limited ability to identify a subset of patients at high risk. DLBCL is highly molecular heterogeneity and there are still a proportion of patients who could not be accurately classified according to the existing molecular classifications. In this study, we found that patients with TP53WT&CD58MUT had the worst outcomes. Interestingly, this subtype of patients harbored enhanced immune escape capacity, giving the abundant infiltration of exhausted T cells and multiple upregulated inhibitory immunomodulatory molecules. Persistent interferon signaling could augment the expression of T-cell inhibitory immune checkpoints such as PD-1, TIM-3, and LAG-3 through JAK/STAT pathway.6 We found that interferon responses and JAK/STAT pathway enriched in the TP53WT&CD58MUT group. Moreover, inhibitory immunomodulatory molecules were preferentially expressed in this group. It suggests that interferon/JAK/STAT pathway-mediated up-regulated expression of inhibitory immunomodulatory molecules could be one potential mechanism through which the inactivation of CD58 facilitated immune evasion and accelerated tumor growth in DLBCL. Despite many of such immune dysregulations had an adverse prognosis; they provided new opportunities for anti-tumor immunotherapy of the subset of DLBCL patients with TP53WT&CD58MUT. Historically, the response rate to anti-PD-1/PD-L1 therapy in unselected DLBCL patients was generally low. Consequently, patients with TP53WT&CD58MUT may be optimal candidates for novel immunotherapy in clinical trials.
In conclusion, our results suggest that TP53 mutation alone is insufficient to effectively differentiate the risk of DLBCL. The mutually exclusive patterns between TP53 and CD58 mutations accurately stratified patients with DLBCL to permit the optional immunotherapy.
ACKNOWLEDGMENTS
This study was supported by Natural Science Foundation of Tianjin grants (19JCYBJC26500 and 18JCZDJC45100), National Natural Science Foundation of China grants (81770213), Clinical Oncology Research Fund of CSCO grants (Y-XD2019-162 and Y-Roche20192-0097), The Science and Technology Research Program of Tianjin Education Commission (2019KJ191), National Key New Drug Creation Special Programs grants (2018ZX09201015), and National Human Genetic Resources Sharing Service Platform/Cancer Biobank of Tianjin Medical University Cancer Institute and Hospital grant (2005DKA21300). The authors thank the Marvel Medical Laboratory, Tianjin Marvelbio Technology Co., Ltd., for providing the assistance in next-generation sequencing and bioinformatics analysis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Xianhuo Wang conceived and designed the study; Xianhuo Wang and Huilai Zhang supervised all aspects of research project and interpreted data; Tingting Zhang, Yaxiao Lu, and Xia Liu performed the research and statistical and bioinformatics analyses; Mengmeng Zhao performed the next-generation sequencing and bioinformatics analysis; Jin He, Xia Liu, Lanfang Li, Lihua Qiu, Zhengzi Qian, and Shiyong Zhou collected samples and clinical information; Bin Meng and Qiongli Zhai reviewed the diagnosis of DLBCL; Xianhuo Wang, Huilai Zhang, and Xiubao Ren provided the clinical samples and material support; Tingting Zhang wrote the manuscript and finalized the figures; Xianhuo Wang. and Huilai Zhang reviewed the manuscript. All authors read and approved the final version of the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
DNA and RNA sequencing data have been submitted to the CNGB Sequence Archive of China National GeneBank DataBase (https://db.cngb.org/cnsa/) under the accession numbers CNP0001322 and CNP0001327, respectively. The mutation data of 1001 DLBCL patients from the Duke University’s cohort were downloaded from cBioPortal (http://www.cbioportal.org/). The mutation and RNA expression data of the patients from the REMoDL-B trail were downloaded from the Supplementary Material from the publication.




