The BCR-ABL1 transcript type influences response and outcome in Philadelphia chromosome-positive chronic myeloid leukemia patients treated frontline with imatinib
Funding information: BolognAIL, AIRC
Abstract
The most frequent BCR-ABL1 fusion transcripts in chronic myeloid leukemia (CML) are the e13a2 (b2a2) and the e14a2 (b3a2) ones. In the imatinib era few studies addressing the prognostic significance of the BCR-ABL1 transcript type in early chronic phase CML have been published. Overall, these studies suggest that in e14a2 patients the response to imatinib is faster and deeper. To evaluate if the BCR-ABL1 transcript type (e13a2 compared to e14a2) affect the response to imatinib and the clinical outcome in newly diagnosed adult CML patients, 559 patients enrolled in 3 prospective studies (NCT00514488, NCT00510926, observational study CML/023) were analyzed. A qualitative PCR was performed at baseline: 52% patients had a e14a2 transcript, 37% a e13a2 transcript, 11% co-expressed both transcripts and 1% had other rare transcripts. The median follow-up was 76 months (95% of the patients had at least a 5-year observation). The complete cytogenetic response rates were comparable in e14a2 and e13a2 patients. The median time to MR3.0 (6 and 12 months) and MR4.0 (41 and 61 months) was significantly shorter for e14a2 patients compared to e13a2 patients, with a higher cumulative probability of MR3.0 (88% and 83%, P < .001) and MR4.0 (67% and 52%, P = .001). The 7-year overall survival (90% and 83%, P = .017), progression-free survival (89% and 81%, P = .005) and failure-free survival (71% and 54%, P < .001) were significantly better in patients with e14a2 transcript. In conclusion, patients with e13a2 transcript had a slower molecular response with inferior response rates to imatinib and a poorer long-term outcome.
1 INTRODUCTION
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by the presence of the BCR-ABL1 fusion gene. The BCR-ABL1 gene encodes a constitutively active tyrosine kinase, which leads to the activation of multiple signaling pathways involved in cell-cycle, adhesion and apoptosis.1, 2 The location of the breakpoints in the ABL1 and BCR genes is variable: the breakpoint in the ABL1 gene can occur anywhere within a large area of more than 300 kb, while in the great majority of CML patients the breakpoints of BCR gene cluster within the Major Breakpoint Cluster Region (M-BCR). The M-BCR (5.8 kb) span exons e12-e16, historically named b1-b5, but the transcribed mRNA has either a e13a2 (b2a2) or a e14a2 (b3a2) junction. In 5-10% of patients, both e13a2 and e14a2 transcripts may be detected by a qualitative RT-QPCR. The fusion protein encoded by BCR-ABL1 varies in size, depending on the breakpoints: both e13a2 and e14a2 mRNAs encode a p210.1-3 In the era of conventional chemotherapy and interferon, several studies have investigated whether the transcript type may influence the outcome of CML patients: overall, these studies failed to detect a significant and robust influence of transcript type on response and clinical outcome.4-7 Imatinib mesylate (IM, Gleevec – Novartis Pharma) is a 2-phenylaminopyrimidine derivative active in Philadelphia-chromosome (Ph) positive CML through a selective inhibition of the BCR-ABL1 protein. In the last decade IM has become the standard therapy of CML patients in early chronic phase (ECP).8-10 In the IM era some studies addressing the prognostic significance of BCR-ABL1 transcript type have been published, suggesting that in e14a2 patients the response to imatinib is faster and deeper.11-14 However, the evidence is weak and mostly limited to response, particularly to molecular response. A recent analysis, conducted within the European Treatment and Outcome Study for CML (EUTOS) CML registry, did not detect survival differences (overall survival and CML-related death) according to the transcript type, but the response and other outcome measures have not been investigated.15 Small differences are difficult to detect, requiring large number of patients and prospective studies. For this purpose, we have analyzed a large cohort of 559 patients who were enrolled in prospective GIMEMA studies of treatment with IM in first-line.
2 METHODS
Five hundred and fifty-nine patients were consecutively enrolled in 3 concurrent multicentric prospective studies of the Italian Group for Hematological Malignancies of Adults (GIMEMA) CML Working Party (WP): CML/021 (Clin Trials Gov. NCT00514488), phase 2, exploring IM 800 mg in intermediate Sokal risk CML patients16; CML/022 (Clin Trials Gov. NCT00510926), phase 3, comparing IM 400 mg vs 800 mg in high Sokal risk CML patients17; CML/023, observational, first-line treatment with IM 400 mg.18 All patients, at least 18 years old, had a Ph+ and/or BCR-ABL1+ CML in ECP (≤ 6 months from diagnosis to IM start; hydroxyurea only allowed). All patients provided written informed consent before the enrollment. The studies were reviewed and approved by the Internal Review Board of all participating institutions, and performed in accordance with the Declaration of Helsinki. A retrospective analysis of the three prospective clinical studies was performed. The intention-to-treat population of each study was analyzed.
The chronic, accelerated or blast disease phase (CP, AP, BP) were defined according to current European LeukemiaNet (ELN) criteria.10 The baseline risk assessment was made using Sokal,19 EURO,20 and EUTOS21 risk scores. Cytogenetic analyses based on G- or Q-banding were performed on bone marrow (BM) samples at baseline, after 6 and 12 months, and every 6 months thereafter, or in case of treatment failure. Cytogenetic responses were defined according to ELN criteria.10 After the achievement of a confirmed complete cytogenetic response (CCyR), if less than 20 metaphases were available, the stability of CCyR could be assessed by fluorescence-in situ-hybridization (FISH) analysis on BM or peripheral blood (PB) cells (CCyR was defined as less than 1% BCR-ABL1 positive nuclei over at least 200 cells).10 A qualitative reverse transcription (RT) polymerase chain reaction (PCR) for BCR-ABL1 transcript was routinely performed at enrollment. Real-time quantitative (RQ) PCR was performed at 3, 6, 12 months, and every 6 months thereafter, according to ELN and EUTOS recommendations.22-26 ABL1 was used as housekeeping gene. A major molecular response (MMR or MR3.0) was defined as a BCR-ABL1 transcript ≤ 0.1%, while a deep molecular response (MR4.0) was defined as a BCR-ABL1 transcript ≤ 0.01% in samples with > 10 000 ABL1 copies.22-26
Baseline characteristics of patients with different transcript types were compared using the Student t test, Pearson's chi square test or Fisher's exact test, as appropriate. The time to response were calculated from treatment start until the first achievement of response. The cumulative incidences of response were estimated under consideration of competing risks27, 28 defined by progression and death. Comparisons between cumulative incidences were performed by the Gray test.29 Overall survival (OS) was calculated from treatment start to death. All deaths, by any cause and at any time (including deaths occurred after IM discontinuation) were considered. Progression-free survival (PFS) was calculated from treatment start to progression or death, whichever came first. Progression was defined as the transformation to AP or BP on IM or subsequent therapy. Failure-free survival (FFS) was calculated from treatment start to IM failure, progression, or death, whichever came first. Failures were retrospectively defined according to 2013 ELN criteria10; as the ELN criteria changed over time, not all the failures according to 2013 ELN criteria were followed by a change of treatment. Probabilities of OS, PFS and FFS were estimated using the Kaplan-Meier method and compared by the log-rank test. A multivariate Cox analysis30 was used to assess the relationship between various predictors of interest and response or outcome. The proportional hazard assumption was verified by Schoenfeld residuals analysis.
3 RESULTS
3.1 Patients
In all the 559 patients a qualitative RT-PCR assay was performed before treatment start: 203 patients (36%) had a e13a2 transcript and 290 patients (52%) had a e14a2 transcript. All analyses and calculations were made on these 493 patients. The remaining patients had both e13a2 and e14a2 transcripts (N = 60, 11%), or other rare transcripts (N = 6, 1%; e1a2 and e19a2 in 2 and 4 patients, respectively). Detailed baseline characteristics according to the transcript type are presented in Table 1. The baseline characteristics of patients expressing both e13a2 and e14a2 transcripts and patients with rare transcripts are presented in Supporting Information Table SI and SII. The patients with e13a2 or e14a2 transcript were comparable for demographic and hematologic characteristics, except for the proportion of male patients, that was higher in the e13a2 group (P = .050), and for the percentages of eosinophils, that was significantly lower in the e13a2 group (P = .009). The patients distribution according to Sokal,19 EURO,20 and EUTOS21 score was comparable. The proportion of patients with clonal chromosomal abnormalities (CCA) in Philadelphia chromosome positive (Ph+) cells at baseline, variant translocations or derivative 9 deletions were similar in the 2 groups. In the e14a2 group, a slightly higher proportion of patients was treated with IM 800 mg daily as initial dose (28%, compared with 20% in the e13a2 group, P = .034).
| Characteristic | e13a2 N = 203 | e14a2 N = 290 | P |
|---|---|---|---|
| Age, years median (range) | 52 (18-79) | 52 (18-84) | .374 |
| Gender Male N (%) | 133 (66) | 164 (57) | .050 |
| ECOG 2, N (%) | 43 (21) | 62 (21) | 1.000 |
| Hb level, g/dL median (range) | 12.0 (7.1-17.2) | 12.3 (6.4-17.5) | .413 |
| PLT count, 103/μL median (range) | 293 (115-4920) | 401 (101-2770) | .251 |
| WBC count, 103/μL median (range) | 61.6 (2.0-500.0) | 52.2 (1.2-491.0) | .174 |
| Peripheral blasts, % median (range) | 1.0 (0-9.5) | 1.0 (0-9.5) | .397 |
| Eosinophils, % median (range) | 1.8 (0-15.0) | 2.0 (0-13.0) | .009 |
| Basophils, % median (range) | 2.0 (0-19.0) | 2.0 (0-16.0) | .259 |
| Spleen, cm median (range) | 1 (0-24) | 1 (0-22) | .693 |
| Sokal score, N (%): | |||
| • Low | 86 (42) | 108 (37) | .525 |
| • Intermediate | 74 (36) | 110 (38) | |
| • High | 43 (21) | 72 (25) | |
| EURO score, N (%): | |||
| • Low | 90 (44) | 124 (43) | .322 |
| • Intermediate | 101 (50) | 142 (49) | |
| • High | 12 (6) | 24 (8) | |
| EUTOS score, N (%): | |||
| • Low | 188 (93) | 267 (92) | .662 |
| • High | 15 (7) | 23 (8) | |
| CCA Ph+ present, N (%) | 7 (3) | 9 (3) | 1.000 |
| Variant translocation present, N (%) | 14 (7) | 13 (4) | .315 |
| Derivative 9 deletions present, N (%) | 27 (13) | 27 (9) | .188 |
| Imatinib dose, N (%): | |||
| • 400 mg | 163 (80) | 208 (72) | .034 |
| • 800 mg | 40 (20) | 82 (28) | |
- Abbreviations: CCA Ph+, clonal chromosome abnormalities in Ph+ cells; ECOG, performance status according to the Eastern Co-operative Oncology Group (all enrolled patients were required to have ECOG performance status 0-2 at baseline); EUTOS, European Treatment and Outcome Study; Hb, hemoglobin; PLT, platelet; WBC, white blood cells.
3.2 Response and outcome by transcript type
The median follow-up was 76 months (range 49-94 months) and 75 months (range 7-99 months) in e13a2 and e14a2 patients, respectively (P = .763). The proportion of patients with an observation equal to or longer than 60 months was similar: 96% and 94% (P =.391) in e13a2 and e14a2 patients, respectively.
The cytogenetic and molecular response rates at milestones are shown in Supporting Information Table SIII: the CCyR rates at 12 months were comparable (75% and 79% in patients with e13a2 or e14a2 mRNA, P = .274), but the rates of MMR at 18 months and MR4.0 at 36 months were significantly lower in patients with e13a2 transcript (52% and 67%, P = .001, and 20% and 30%, P = .013, respectively). The proportion of not evaluable patients was comparable in the 2 groups.
As far as the rapidity of response, the median time to CCyR was 6 months in both groups, with a comparable overall estimated probability of CCyR: 89% and 88% in patients with e13a2 and e14a2 transcript, P = .916. The patients with e13a2 transcript achieved a MMR and a MR4.0 significantly slower than patients with e14a2 transcript: the median time to MMR was 12 and 6 months, with 83% and 88% overall estimated probability of MMR in e13a2 patients and e14a2 patients (P < .001), respectively; the median time to MR4.0 was 61 and 41 months and the overall estimated probability of MR4.0 was 52% and 67% in the 2 groups (P = .001), respectively (Figure 1).
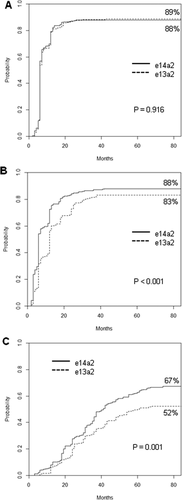
Time to response by transcript type. Estimates of (A) time to complete cytogenetic response, (B) time to major molecular response, and (C) time to deep molecular response (MR4.0), according to the fusion transcript type (patients included: 203 patients with e13a2 transcript and 290 patients with e14a2 transcript)
The 7-year estimated probabilities of OS (83% and 90%, P = .017), PFS (81% and 89%, P = .005) and FFS (54% and 71%, P < .001) were significantly lower in patients with e13a2 transcript (Figure 2). In multivariate Cox analysis,30 the transcript type retained its prognostic significance on time to MMR, time to MR4.0, OS, PFS and FFS (Supporting Information Table SIV), when adjusted for other relevant variables.
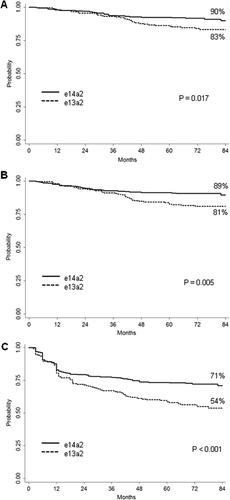
Outcome by transcript type. Estimates of (A) overall survival, (B) progression-free survival, (C) failure-free survival, according to the fusion transcript type (patients included: 203 patients with e13a2 transcript and 290 patients with e14a2 transcript). The definitions of progression and failure are detailed in the methods section
The patients receiving a salvage therapy (one or more tyrosine kinase inhibitors, conventional chemotherapy, allogeneic stem cell transplantation) were 64 (31%) and 71 (24%) in the e13a2 and e14a2 groups, respectively. Molecular data after treatment change were not prospectively collected, so the impact on molecular response cannot be assessed. The patients who died after progression to advanced phase were 13/64 (20%) and 13/71 (18%), respectively.
The responses and the survival probabilities of patients co-expressing the e13a2 and the e14a2 transcripts were similar to or even better than the ones of e14a2 patients (Supporting Information Figures S1 and S2). The response and the outcome of patients with rare transcripts are presented in Supporting Information Table SII.
3.3 Prognostic impact of transcript type according to the initial imatinib dose
Analyzing separately the patients treated with IM 400 mg (n = 371; e13a2 and e14a2 transcript in 163 and 208 patients, respectively) or IM 800 mg daily (n = 122; e13a2 and e14a2 transcript in 40 and 82 patients, respectively), the response differences according to the transcript type (e13a2 vs e14a2) were confirmed: the cumulative incidences of MMR were 84% and 89% in patients treated with standard-dose IM (median time to MMR: 12 and 6 months, respectively, P = .001) and 73% and 82% in patients treated with high-dose IM (median time to MMR: 15 and 6 months, respectively, P = .026); the cumulative incidences of MR4.0 were 57% and 68% in patients treated with 400 mg (median time to MR4.0: 54 and 41 months, respectively, P = .024) and 30% and 60% in patients treated with 800 mg daily (median time to MR4.0 not reached and 48 months, respectively, P = .007).
In patients treated with standard-dose IM, the transcript type was able to predict significantly different 7-year probabilities of OS, PFS and FFS (Figure 3). In the high-dose IM group, the patients with e13a2 transcript had inferior PFS (71% and 86%, P = .039) and FFS (44% and 67%, P = .011), but no difference could be detected in OS (82% and 87%, P = .232) (Figure 3).
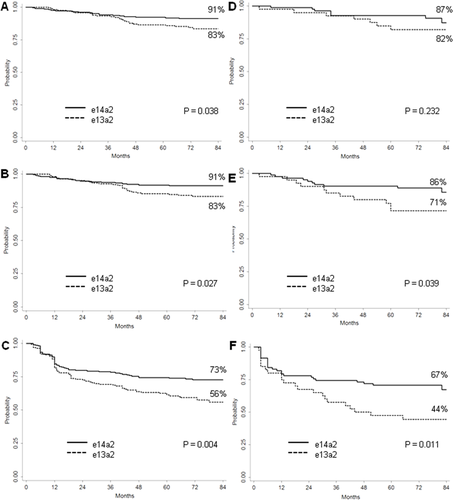
Outcome by transcript type in patients treated with standard- or high-dose imatinib. Estimates of (A) overall survival, (B) progression-free survival, (C) failure-free survival, according to the fusion transcript type in patients receiving standard-dose imatinib (patients included: 163 patients with b2a2 transcript and 208 patients with b3a2 transcript). Estimates of (D) overall survival, (E) progression-free survival, (F) failure-free survival, according to the fusion transcript type in patients receiving high-dose imatinib (patients included: 40 patients with b2a2 transcript and 82 patients with b3a2 transcript; all patients treated with high-dose imatinib had intermediate or high Sokal score). The definitions of progression and failure are detailed in the methods section
4 DISCUSSION
The great majority of CML patients have a mRNA transcript with a e13a2 or a e14a2 junction.1-3 The information on the prognostic impact of BCR-ABL1 transcript type in ECP CML patients treated frontline with IM is still limited to few studies. In 74 patients treated with IM 400 mg it was found that the CCyR rate at one year was higher in e14a2 patients than in e13a2 ones (54% vs 24%, P = .01).11 In a much larger study of 1105 patients, it was found that the cumulative probability of achieving MMR and MR4.0 was significantly higher in e14a2 patients than in e13a2 ones.12 In 481 patients treated frontline with IM or with second generation TKIs, the CCyR and the MMR rate at several time points were higher in e14a2 patients.13 A MR4.0 was reported more frequently in e14a2 patients than in e13a2 ones (57% vs 27%, P = .003).14 However, in only one study13 a difference in outcome (transformation-free survival, 98% versus 91%, P = .01) was detectable; no differences in overall survival or other outcome measures were reported. A large analysis on 1494 European patients failed to detect survival differences, considering either deaths for any reason or CML-related deaths; responses and other outcome measures have not been analyzed.15
In line with prior reports, we have also found that the e13a2 transcript was associated with a poorer response to IM: MMR and MR4.0 rates were significantly lower with a significantly longer time to the first MMR and MR4.0. As predictable, since CCyR and MMR are the best surrogate markers of the outcome,8-10 we also found that the e13a2 transcript was associated with a poorer outcome (OS, PFS and FFS). Moreover, the deep molecular response may predict the probability of achieving a treatment-free remission (TFR).10 The differences between transcripts were not influenced by confounding factors: the analysis was performed considering the intention-to-treat population of each protocol, the proportion of patients lacking of cytogenetic or molecular evaluation (weighting as a negative result) was balanced between the two groups, and the relative risk distribution according to Sokal,19 EURO,20 and EUTOS21 scoring systems was comparable. By multivariable analysis, the prognostic value of transcript type was even stronger than that of any other variables.
There were some differences between the two groups. In the e13a2 group there were more males, and males may have a worse prognosis; moreover, in the same group the baseline percentage of eosinophils in peripheral blood (PB) was lower, and eosinophils have been shown to have a worse prognosis.20 More importantly, the proportion of patients treated with IM 800 mg daily as initial dose was lower in e13a2 group. High-dose IM (equal to or higher than 600 mg daily) has not been approved yet as first-line therapy of CML patients in CP. Four randomized studies comparing 400 mg versus 800 mg IM daily have been published, three for patients of any risk category31-33 and one for high Sokal score patients only.17 A significant advantage of high-dose treatment was shown in only one study,32 but in all four studies there was a trend in favor of high doses. In our study, due to the inclusion and exclusion criteria of studies CML/021 and CML/022, all the patients treated with high dose IM had intermediate or high Sokal score. To test if IM dose could have influenced the results of this study, we made all calculations separately, according to the initial IM dose, and we found that in e13a2 patients responses and outcomes were poorer irrespective of IM dose, with the exception of OS in patients treated with high-dose (no significant difference).
The difference between e13a2 and e14a2 patients is difficult to understand and to explain. The e13a2 and e14a2 hybrid mRNA and protein differ in size by 75 bases and 25 amino acids, respectively. These sequences are a putative site for calcium-dependent lipid binding, but the exact function is currently unknown.1-3 With conventional chemotherapy or alpha-interferon, the BCR-ABL1 transcript type had no strong effects on the response and the outcome.4-7 Considering the selectivity of IM, the emergence of outcome differences between e13a2 and e14a2 patients suggests a potential BCR-ABL1 dependent mechanism. A higher pCrKL/CrKl ratio at diagnosis has been described in patients with a e13a2 transcript,11 but the higher pre-treatment tyrosine kinase activity was not evaluated in a multivariate analysis including other relevant variables (ie, disease score). We may hypothesize a more efficient tyrosine kinase activity in e13a2 patients, but a systematic evaluation of the pre-treatment tyrosine kinase activity has not been accomplished in our study. Conversely, the junction sequences product of the BCR-ABL1 fusion can bind several human leukocyte antigen (HLA) class I and II molecules, and to elicit peptide-specific T-cell responses.34-38 We may speculate on a putative different immunogenic ability of the e13a2 protein, which is shorter than the e14a2 one and have different junctional sequences, but no evidences supporting this hypothesis are available. Finally, the response and outcome differences between e13a2 and e14a2 patients could be due to the crystallographic structure of the BCR-ABL protein or to a different interaction between the IM molecule and the p210 protein, but clarifying data have not been published yet.
The current CML therapeutic scenario has been enriched by FDA and EMA approval of nilotinib (NIL) and dasatinib (DAS) as frontline treatment. In early CP CML, both drugs induced significantly superior rates of cytogenetic and molecular responses as compared with IM.39-42 So far, few data are available about the relationship between the BCR-ABL1 transcript type and the response to second generation TKIs: a preliminary study from the GIMEMA CML WP on 345 patients treated with NIL based regimens, despite a trend for lower response rates and inferior outcome in patients with e13a2 transcript, did not detect significant differences between e13a2 and e14a2 patients.43 Another preliminary study from MD Anderson Cancer Center on 204 patients treated with NIL or DAS, reported a lower rate of molecular responses and a trend for inferior EFS in e13a2 patients.44
We conclude that the e13a2 BCR-ABL1 fusion transcript affects the rate, the depth, and the speed of the response to treatment with imatinib firstline, and that including the transcript type in the calculation of the baseline risk scores may improve prognostic stratification and may help the choice of the best treatment policy. More data and a longer follow-up, are required to understand if the transcript type influences the immunologic control of residual disease, so affecting the probability of achieving a TFR.
ACKNOWLEDGMENTS
This study has been supported by: BolognAIL and AIRC.
The following members of the “GIMEMA Working Party on CML,” formerly “ICSG on CML” actively participated in this study, enrolling patients and collecting clinical data: Lucarelli G, Polimeno G (Internal Medicine Unit, “F. Miulli” Hospital, Acquaviva delle Fonti, Bari); Ladetto M, Pini M (Department of Hematology, A.O. Nazionale Santi Antonio e Biagio e C. Arrigo, Alessandria); Scortechini AR, Leoni P (Hematology Department, University of Ancona, Azienda Ospedaliero Universitaria Ospedali Riuniti di Ancona, Ancona); Galieni P, Bigazzi C (Hematology Unit, Presidio Ospedaliero “C. e G. Mazzoni,” Ascoli Piceno); Cantore N, Palmieri F (Hematology Division, Ospedale Civile “San Giuseppe Moscati,” Avellino); Specchia G, Russo Rossi A (Chair of Hematology, University of Bari, Bari); Rambaldi A, Intermesoli T (Hematology Unit, Azienda Ospedaliera Papa Giovanni XXIII, Bergamo); Palandri F, Luatti S, Testoni N, Iacobucci I, Venturi C, Mancini M, Apolinari M, Fogli M (Institute of Hematology “Seràgnoli,” Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Bologna); Capucci A, D'Adda M (Hematology Unit, Azienda Ospedaliera “Spedali Civili,” Brescia); Melpignano A, Girasoli M (Hematology Division, Ospedale “Perrino,” Brindisi); Cabras MG (Hematology Unit, Ospedale Oncologico “A. Businco,” Cagliari); De Biasi E (Hematology Unit, Presidio Ospedaliero Camposampiero, Camposampiero, Padova); Tagariello G, Sartori R (Hematology Unit, “San Giacomo” Hospital, Castelfranco Veneto, Treviso); Di Raimondo F, Stagno F (Hematology Unit, “Ferrarotto” Hospital, Catania); Molica S, Lentini M (Hematology Unit, “Pugliese” Hospital, Catanzaro); Spedini P, Viganò C (Hematology and BMT Unit, “Istituti Ospitalieri di Cremona” Hospital, Cremona); Massaia M, Rapezzi D (Division of Hematology, “Santa Croce e Carle” Hospital, Cuneo); Cuneo A, Ciccone M (Chair of Hematology, Dipartimento di Scienze Mediche, “Arcispedale S. Anna” University Hospital, Ferrara); Bosi A, Gozzini A (Chair of Hematology, University of Firenze, Firenze); Gobbi M, Bergamaschi M (Chair of Hematology, IRCCS San Martino, Genova); Chianese R (Hematology Unit, Ospedali Riuniti ASL TO4, Ivrea, Torino); Cimino G, Ciccone F (Hematology Unit, Ospedale Civile, Latina); Capochiani E, Pelosini M (Oncology and Hematology Unit, Ospedali Civili, Livorno; Musolino C, Russo S (Division of Hematology, University of Messina, Messina); Cortelezzi A (Hematology Unit, Oncohematology Division, IRCCS Ca' Granda – Maggiore Policlinico Hospital Foundation, Milano); Luppi M, Marasca R (Chair of Hematology, University of Modena and Reggio Emilia, Modena); Gambacorti-Passerini C (Department of Hematology, “San Gerardo” Hospital, Monza); Luciano L, Izzo B (Department of Biochemistry and Medical Biotechnologies, “Federico II” University, Napoli); Ferrara F (Hematology and Bone Marrow Transplantation Unit, “Cardarelli” Hospital, Napoli); Mettivier V, Sessa U (Hematology Unit, “Cardarelli” Hospital, Napoli); Latte G, Noli D (Hematology Unit, “San Francesco” Hospital, Nuoro); Rege-Cambrin G (Department of Clinical and Biological Sciences, “San Luigi Gonzaga” University Hospital, Orbassano, TO); Semenzato G, Binotto G (Department of Internal Medicine, University of Padova, Padova); Fabbiano F, Malato A (Hematology Unit, “V. Cervello” Hospital, Palermo); Siragusa S, Caracciolo C (Chair of Hematology, University of Palermo, Palermo); Musso M, Porretto F (Oncology and Bone Marrow Transplantation Unit, “La Maddalena” Hospital, Palermo); Cazzola M (Hematology Unit, “S. Matteo” University Hospital, Pavia); Falini B, Falzetti F (Division of Hematology and Clinical Immunology, Department of Medicine, University of Perugia, Perugia); Visani G, Isidori A (Hematology Unit, “San Salvatore” Hospital, Pesaro); Di Bartolomeo P, Di Lorenzo R (Hematology Unit, Ospedale Civile dello Spirito Santo, Pescara); Vallisa D, Trabacchi E (Hematology Division, “Guglielmo da Saliceto” Hospital, Piacenza); Pizzuti M (Hematology Unit, “San Carlo” Hospital, Potenza); Lanza F, Salvucci M (Hematology Unit, “Santa Maria delle Croci” Hospital, Ravenna); Ronco F, Ielo D (Hematology Unit, Ospedali Riuniti, Reggio Calabria); Merli F, Capodanno I (Hematology Unit, Arcispedale Santa Maria Nuova, Reggio Emilia); Tosi P, Merli A (Hematology Unit, Ospedale Infermi Azienda Unità Sanitaria, Rimini); Sorà F, (Istituto di Semeiotica Medica, Università Cattolica del Sacro Cuore-Policlinico “Gemelli,” Roma); Latagliata R, Foà R (Chair of Hematology, “La Sapienza” University, Roma); De Fabritiis P, Trawiska M (Hematology Unit, “S. Eugenio” Hospital, Roma); Amadori S, Cantonetti M (Department of Hematology, “Tor Vergata” University, Roma); Pierelli L, Pacilli L (Hematology and Stem Cell Transplantation Unit, Azienda Ospedaliera San Camillo Forlanini, Roma); Bagnato A, Cedrone M (Hematology Unit, Ente Ospedaliero San Giovanni Addolorata, Roma); Mengarelli A, Romano A (Hematology Unit, Istituto Regina Elena, Roma); Tafuri A, Montefusco E (Hematology Unit, “Sant'Andrea” Hospital, Roma); Iuliano F, Infusino S (Hematology Unit, Presidio Ospedaliero “N. Giannettasio,” Rossano Calabro); Dore F, Fozza C (Institute of Hematology, University of Sassari, Sassari); Aprile L, Defina M (Chair of Hematology, University of Siena, Siena); Liberati AM, Luzi D (Hematology and Oncology Unit, Azienda Ospedaliera “S. Maria,” Terni); Boccadoro M, Ferrero D (Section of Hematology, Department of Molecular Biotechnology and Health Sciences, University of Torino, Torino) Vitolo U, Nicolosi M (Hematology Unit, Azienda Ospedaliero Universitaria Città della Salute e della Scienza, University of Torino, Torino); Gottardi M, Calistri E (Hematology Unit, “Ca' Foncello” Hospital, Treviso); Fanin R (Chair of Hematology, University of Udine, Udine); Pizzolo G, Krampera M (Chair of Hematology, University of Verona, Verona); Di Bona E (Hematology Unit, Ospedale Civile, Vicenza).
CONFLICT OF INTERESTS
F. Castagnetti: consultant for and honoraria from Novartis, Bristol-Myers Squibb, Pfizer and Incyte; G. Gugliotta: consultant for and honoraria from Novartis and Bristol-Myers Squibb; M. Breccia: consultant for Bristol-Myers Squibb, Novartis, Pfizer and Incyte; A. Iurlo: consultant for and honoraria from Novartis, Bristol-Myers Squibb and Pfizer; P Vigneri: consultant for and honoraria from Incyte, Bristol-Myers Squibb and Novartis, honoraria from Pfizer; E. Abruzzese: consultant for Novartis and Bristol-Myers Squibb; M. Tiribelli: consultant for and honoraria from Novartis, Bristol-Myers Squibb and Incyte; S. Soverini: consultant for and honoraria from Novartis, Bristol Myers Squibb and Incyte; M. Cavo: consultant for and honoraria from Celgene, Janssen, Takeda, Amgen and Bristol Myers Squibb; G. Martinelli: consultant for and honoraria from Incyte, Bristol-Myers Squibb, Novartis and Pfizer; G. Saglio: consultant for and honoraria from Bristol-Myers Squibb, Novartis, Incyte and Celgene; F. Pane: research support from Novartis, consultant for Novartis, Bristol-Myers Squibb and Incyte, honoraria from Novartis and Bristol-Myers Squibb; M. Baccarani: consultant for and honoraria from Novartis, Bristol-Myers Squibb, Pfizer and Incyte; G. Rosti: consultant for and honoraria from Novartis, Bristol Myers Squibb, Incyte, Pfizer and Roche; the remaining Authors had not relevant conflicts of interest to disclose.




