DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia
Abstract
DNMT3A mutations are seen in ∼5% of patients with chronic myelomonocytic leukemia (CMML) and thus far, have had an indeterminate prognostic impact on survival. We carried out this study to assess the prognostic impact of DNMT3A mutations on a larger informative cohort of CMML patients (n = 261). DNMT3A mutations were seen in 6% (n = 16); 56% (n = 9) male, with a median age of 64 years. Eighty-one % of DNMT3A mutations were missense, with the Arg882 mutational hot spot accounting for 63% of all changes. Five (31%) patients had an abnormal karyotype whereas concurrent gene mutations (SF3B1/SRSF2/U2AF1−56%, TET2−50%, and ASXL1−25%) were seen in all patients. Apart from a higher frequency of SF3B1 (P = 0.0001) and PTPN11 (P = 0.005) mutations and a lower frequency of SRSF2 (P = 0.004) mutations, there were no significant differences between DNMT3A mutated patients and their wildtype counterparts. In univariate analysis, survival was shorter in DNMT3A mutated (median 8 months) versus wildtype (median 27 months) patients (P = 0.0007; HR 2.9, 95% CI 1.5–5.7); with other variables of significance including lower hemoglobin (P = 0.002), higher leukocyte count (P = 0.0009), higher monocyte count (P = 0.0012), circulating blast % (P = 0.001), circulating immature myeloid cells (P = 0.01), bone marrow blast % (P = 0.045), abnormal karyotype (P = 0.02), and ASXL1 (P = 0.01) mutations. In a multivariable model that included the aforementioned variables, when both DNMT3A and ASXL1 mutations were added, only DNMT3A (P < 0.0001) and ASXL1 (P = 0.004) mutations remained significant. DNMT3A mutations were also predictive of a shortened leukemia-free survival. These findings warrant inclusion of DNMT3A mutations in molecularly integrated CMML prognostic models. Am. J. Hematol. 92:56–61, 2017. © 2016 Wiley Periodicals, Inc.
Introduction
Chronic myelomonocytic leukemia (CMML) is a clonal hematopoietic stem cell disorder with overlapping features of myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN) 1. Clonal cytogenetic abnormalities are seen in ∼20–30% of patients, whereas gene mutations are seen in >90% 2-5. Common mutations include, TET2 (∼60%), SRSF2 (∼45%), ASXL1 (∼40%), and RAS (∼30%) 4, 6. Of these, thus far, only frameshift and nonsense ASXL1 mutations have been shown to independently, adversely impact survival 2, 6. This has led to the incorporation of ASXLI mutations in molecularly integrated CMML prognostic models including the Groupe Français des Myélodysplasies (GFM) model, Mayo Molecular Model (MMM) and the CMML specific prognostic scoring system- molecular (CPSS-Mol) 2, 6, 7.
DNA methylation is mediated by a family of DNA methyltransferase enzymes (DNMT), including DNMT1, DNMT3A (chromosome 2p23), and DNMT3B 8. DNMT1 primarily maintains pre-existing DNA methylation patterns, whereas DNMT3A and DNMT3B carry out de novo DNA methylation 8. DNMT3A mutations are common in hematological malignancies and occur in ∼20% of patients with acute myeloid leukemia (AML), ∼13% with MDS, ∼10% with MPN, 2–5% with CMML and ∼20% with T-cell lymphoproliferative disorders; with some, but not all studies demonstrating an adverse impact on survival 3, 9-12. Of note, a recurrent Arginine882 (R882) hot spot accounts for 40–60% of DNMT3A mutations, with limited data suggesting loss of methyltransferase activity in in vitro assays 13.
In CMML, DNMT3A mutations are seen in ∼2–5% of patients. In a large GFM study (n = 312), DNMT3A mutations were seen in 2% and were not included in further survival analyses 6. In a prior Mayo Clinic study (n = 175), DNMT3A mutations were seen in 5% (n = 9) and on univariate, but not multivariate analysis, were associated with shortened overall survival (OS) 3. We carried out this study on a larger CMML cohort (n = 261), with more (n = 16) informative cases to assess the impact of DNMT3A mutations in patients with CMML.
Materials and Methods
Two-hundred and sixty-one patients with CMML were included in the study. All patients had bone marrow (BM) biopsies and cytogenetic studies performed at diagnosis. The diagnosis of CMML, including subclassification into CMML-0, CMML-1 and CMML-2, and leukemic transformation (LT) were according to the 2008 World Health Organization (WHO) criteria and the recent 2016 WHO revision recommendations 14, 15. Risk stratification was per the Mayo-French cytogenetic system 5, Mayo prognostic model 16, GFM model 6, and the Mayo Molecular Model 2. Twenty-eight gene panel targeted capture assays were carried out on BM DNA specimens obtained at diagnosis for the following genes; TET2, DNMT3A, IDH1, IDH2, ASXL1, EZH2, SUZ12, SRSF2, SF3B1, ZRSR2, U2AF1, PTPN11, Tp53, SH2B3, RUNX1, CBL, NRAS, KRAS, JAK2, CSF3R, FLT3, KIT, CALR, MPL, NPM1, CEBPA, IKZF, and SETBP1, by previously described methods 3.
The flow cells were sequenced as 101 X 2 paired end reads on an Illumina HiSeq 2000 using TruSeq SBS sequencing kit version 3 and HiSeq data collection version 2.0.12.0 software. Base-calling was performed using Illumina's RTA version 1.17.21.3. Genesifter® software was utilized (PerkinElmer, Danvers, MA) to analyze targeted sequence data. Reads from the sequencing in fastq format were aligned using Burrows–Wheeler Aligner (BWA) against the genomic reference sequence for Homo sapiens (Build 37.2; NCBI http://www.ncbi.nlm.nih.gov/). An additional alignment, postprocessing set of tools was then used to do local realignment, duplicate marking, and score recalibration to generate a final genomic aligned set of reads. Nucleotide variants were called using the Genome Analysis Toolkit (GATK-Broad Institute, Cambridge, MA), which identified single nucleotide and small insertion/deletion events using default settings. Specific variants were deemed as mutations if they are associated with a heme malignancy (as identified by COSMIC database), or if they have not been associated with a dbSNP.
Based on prior observations, only frame shift and nonsense ASXL1 mutations were considered pathogenic 2, 16. For TET2, frame shift, nonsense, missense, insertions, and deletions were considered pathogenic 17. Previously annotated single nucleotide polymorphisms (http://www.hapmap.org) in all the aforementioned genes, were considered nonpathogenic.
All statistical analyses considered parameters obtained at time of referral to the Mayo Clinic, which in most instances coincided with time of BM biopsy. Differences in the distribution of continuous variables between categories were analyzed by either Mann–Whitney (for comparison of two groups) or Kruskal–Wallis (comparison of three or more groups) test. Patient groups with nominal variables were compared by chi-square test. Overall survival was calculated from the date of first referral to date of death (uncensored) or last contact (censored). Leukemia-free survival (LFS) was calculated from the date of first referral to date of leukemic transformation (uncensored) or death/last contact (censored). Overall and LFS curves were prepared by the Kaplan–Meier method and compared by the log-rank test. Cox proportional hazard regression model was used for multivariable analysis. P values less than 0.05 were considered significant. The Stat View (SAS Institute, Cary, NC) statistical package was used for all calculations.
Results
Among the 261 study patients, 64% (n = 168) were males and median age was 70 years (range, 20–91). One-hundred and fifty four (59%), 65 (25%), and 42 (16%) patients were classified as CMML-0, 1 and 2, respectively. Table 1 describes the clinical and laboratory features and subsequent events in 261 patients with WHO defined CMML, stratified by the presence or absence of DNMT3A mutations. At a median follow-up of 23 months, 174 (67%) deaths and 37 (14%) leukemic transformations (LT) were documented. Eighty-four (32%) patients had an abnormal karyotype, of which, 10 (4%) were complex and 3 (1%) were monosomal. Risk stratification based on the Mayo-French cytogenetic system included: 180 (72%) low risk, 57 (23%) intermediate risk, and 13 (5%) high risk, respectively. Mutational frequencies included; TET2 (n = 118) 45%, ASXL1 (n = 120) 45%, SRSF2 (n = 105) 40%, NRAS (n = 37)14%, SETBP1 (n = 34) 13%, CBL (n = 25) 10%, JAK2V617F (n = 17) 7%, RUNX1 (n = 16) 6%, DNMT3A (n = 16) 6%, U2AF1 (n = 16) 6%, SF3B1 (n = 13) 5%, ZRSR2 (n = 5) 4%, Tp53 (n = 10) 4%, IDH2 (n = 11) 4%, KRAS (n = 8) 3%, c-KIT (n = 7) 3%, PTPN11 (n = 6) 2%, IDH1 (n = 4) 2%, and ≤1% for EZH2 (n = 3), CSF3R (n = 3), CALR (n = 1), MPL (n = 1) and SH2B3 (n = 1), respectively. There were no mutations seen in IKZF, NPM1, and PHF6. Risk stratification by the Mayo Molecular Model included: 26 (10%) low risk, 72 (28%) intermediate 1, 79 (30%) intermediate 2, and 81 (31%) high risk, respectively.
| Variable | All patients with CMML (n = 261) | CMML patients with DNMT3A mutations (n = 16) |
CMML patients without DNMT3A mutations (n = 245) |
P value |
|---|---|---|---|---|
| Age in years; median (range) | 70 (20–91) | 65 (20–87) | 70 (27–91) | 0.07 |
| Males; n (%) | 168 (64) | 9 (56) | 159 (65) | 0.48 |
| Hemoglobin g/dL; median (range) | 10.6 (6.4–16.9) | 9.6 (6.8–13.2) | 10.7 (6.4–16.9) | 0.09 |
| MCV femtoliter; median (range) | 91 (59–119) | 91 (75–112) | 91 (59–119) | 0.5 |
| WBC × 109/L; median (range) | 12.1 (1.5–264) | 12.6 (2.9–71.5) | 12 (1.5–264) | 0.83 |
| ANC × 109/L; median (range) | 5.8 (0–151) | 6.7 (1–39.2) | 5.7 (0–151) | 0.74 |
| AMC × 109/L; median (range) | 2.3 (1.0–40) | 1.7 (1.0–20) | 2.4 (1.0–40) | 0.76 |
| ALC × 109/L; median (range) | 1.7 (0–22) | 1.9 (0.4–5.6) | 1.7 (0–22) | 0.82 |
| Platelets × 109/L; median (range) | 97 (10–840) | 112 (11–840) | 96 (10–726) | 0.45 |
| Presence of circulating immature myeloid cells; n (%) | 142 (54) | 9 (60) | 133 (55) | 0.7 |
| PB blast %; median (range) | 0 (0–19) | 0 (0–19) | 0 (0–7) | 0.3 |
| BM blast %; median (range) | 3 (0–19) | 3 (0–13) | 3 (0–19) | 0.9 |
| BM cellularity (%) | 80 (40–100) | |||
| Lactate dehydrogenase levels IU/mL; n (range) | 225 (84–1296) | 223 (109–294) | 225 (84–1296) | 0.48 |
| Next generation sequencing analysis; n (%) | ||||
| 1. Epigenetic regulators | ||||
| TET2 | 118 (45) | 8 (50) | 110 (45) | 0.7 |
| IDH1 | 4 (2) | 0 (0) | 4 (2) | 0.6 |
| IDH2 | 11 (4) | 0 (0) | 11 (4) | 0.38 |
| 2. Chromatin regulation | ||||
| ASXL1 | 120 (50) | 6 (37) | 114 (51) | 0.3 |
| EZH2 | 3 (1) | 0 (0) | 3 (1) | 0.656 |
| SUZ12 | 0 | 0 (0) | 0 (0) | – |
| 3. Transcription factors | ||||
| RUNX1 | 16 (6) | 2 (12) | 14 (6) | 0.27 |
| 4. Spliceosome components | ||||
| SF3B1 | 14 (5) | 5 (25) | 9 (4) | 0.0001 |
| SRSF2 | 105 (40) | 1 (6) | 104 (42) | 0.004 |
| U2AF1 | 16 (6) | 2 (12) | 14 (6) | 0.2 |
| ZRSR2 | 5 (3) | 0 (0) | 5 (2) | 0.8 |
| 5. Cell signaling | ||||
| JAK2 V617F | 17 (7) | 1 (6) | 16 (7) | 0.9 |
| CALR | 1 (0.5) | 0 (0) | 1 (0.5) | 0.8 |
| MPL | 1 (0.4) | 0 (0) | 1 (0.5) | 0.8 |
| SH2B3 | 1 (0.5) | 0 (0) | 1 (0.5) | 0.8 |
| CBL | 25 (10) | 0 (0) | 25 (10) | 0.4 |
| KRAS | 8 (3) | 0 (0) | 8 (3) | 0.5 |
| NRAS | 37 (14) | 2 (16) | 35 (14) | 0.8 |
| PTPN11 | 6 (2) | 2 (12) | 4 (2) | 0.005 |
| CSF3R | 3 (1) | 0 (0) | 3 (1) | 0.7 |
| C-KIT | 7 (3) | 1 (6) | 6 (2) | 0.4 |
| FLT3TKD | 1 (0.5) | 0 (0) | 1 (0.5) | 0.8 |
| NPM1 | 0 | 0 (0) | 0 (0) | – |
| 6. Tumor suppressor genes | ||||
| Tp53 | 10 (4) | 2 (12) | 9 (4) | 0.09 |
| PHF6 | 0 | 0 (0) | 0 (0) | – |
| 7. Others | ||||
| SETBP1 | 34 (13) | 2 (12) | 32 (13) | 0.9 |
| IKZF | 0 | 0 (0) | 0 (0) | – |
| 2008 WHO morphological subtypes; n (%) | ||||
| CMML-1 | 221 (84) | 13 (81) | 208 (85) | 0.7 |
| CMML-2 | 40 (16) | 3 (19) | 37 (15) | |
| 2016 WHO morphological subtypes; n (%) | ||||
| CMML-0 | 154 (59) | 10 (62) | 144 (59) | 0.9 |
| CMML-1 | 65 (25) | 4 (25) | 61 (25) | |
| CMML-2 | 42 (16) | 2 (12) | 40 (16) | |
| Spanish Cytogenetic risk stratification; n (%)a | ||||
| Low | 180 (72) | 11 (69) | 169 (72) | 0.1 |
| Intermediate | 43 (17) | 1 (6) | 42 (18) | |
| High | 27 (11) | 4 (25) | 23 (10) | |
| Mayo-French cytogenetic risk stratification; n (%)a | ||||
| Low | 180 (72) | 11 (69) | 169 (72) | 0.4 |
| Intermediate | 57 (23) | 3 (19) | 54 (23) | |
| High | 13 (5) | 2 (12) | 11 (5) | |
| Mayo prognostic model; n (%) | ||||
| Low | 89 (34) | 3 (20) | 86 (35) | 0.2 |
| Intermediate | 83 (32) | 4 (27) | 79 (32) | |
| High | 87 (34) | 8 (53) | 79 (32) | |
| Mayo Molecular Model; n (%) | ||||
| Low | 26 (10) | 0 (0) | 26 (11) | 0.5 |
| Intermediate 1 | 72 (28) | 4 (25) | 68 (28) | |
| Intermediate 2 | 79 (30) | 6 (37) | 73 (30) | |
| High | 81 (31) | 6 (37) | 75 (31) | |
| GFM CMML prognostic model; n (%) | 0.4 | |||
| Low | 119 (46) | 9 (56) | 110 (45) | |
| Intermediate | 92 (36) | 6 (37) | 86 (35) | |
| High | 48 (18) | 1 (6) | 47 (19) | |
| Leukemic transformations; n (%) | 37 (14) | 4 (25) | 33 (13) | 0.2 |
| Deaths; n (%) | 174 (67) | 10 (62) | 164 (67) | 0.7 |
- CMML, chronic myelomonocytic leukemia; WHO, World Health Organization; FAB, French American British; MCV, mean corpuscular volume; WBC, white blood cell count; ANC, absolute neutrophil count; AMC, absolute monocyte count; ALC, absolute lymphocyte count; PB, peripheral blood; BM, bone marrow, GFM, Groupe Français des Myélodysplasies.
- a Cytogenetic studies were available for 250 patients with chronic myelomonocytic leukemia at diagnosis.
DNTM3A mutated CMML: phenotypic and molecular correlates
DNMT3A mutations were seen in 16 (6%) patients; 56% (n = 9) male with a median age of 64 years (range, 27–91). Five (31%) patients had an abnormal karyotype; including 2 (12%) with a complex karyotype and 4 (25%) with chromosome 7 abnormalities (monosomy7/del7q). Table 2 describes the clinical and laboratory characteristics of 16 patients with WHO defined, DNMT3A mutated CMML. DNMT3A amino acid substitutions included; Arg882His (n = 7) 44%, Arg882Cys (n = 3) 19%, and 6% (n = 1) each for Ala910Pro, Arg598*, Arg320*, Val636Ala, and Leu859Tyr, respectively (Fig. 1). Of note, 1 (6%) patient had an intronic 5′ splice site mutation that was predicted to result in loss of the splice site donor sequence (by in silico analysis). In 13 (81%) patients DNMT3A mutations produced changes affecting the catalytic methyltransferase domain, whereas in 1 (6%) each, mutations affected the ADD (ATRX-DNMT3-DNMT3L) and PWWP (Pro-Trp-Trp-Pro) domains, respectively (Fig. 1A). In comparison to other myeloid neoplasms such as MDS and MDS/MPN overlap syndromes with ring sideroblasts and thrombocytosis, there were no mutations affecting the N-terminal region in CMML patients (Fig. 1B). There were 13 (81%) missense mutations and 2 (12%) nonsense mutations; with a median mutant allele frequency burden of 45% (individual values in Table 2). Concurrent gene mutations were detected in; TET2 (n = 8) 50%, ASXL1 (n = 4) 25%, SF3B1 (n = 5) 31%, SRSF2 (n = 2) 12.5%, U2AF1 (n = 2) 12.5%, PTPN11 (n = 2) 6%, RUNX1 (n = 2) 12.5%, SETBP1 (n = 2) 12.5%, Tp53 (n = 2) NRAS (n = 1) 6%, c-KIT (n = 1) 6%, and CBL (n = 1) 6%, respectively. Cumulatively 56% had one or more spliceosome component mutations (SF3B1, SRSF2, and U2AF1). Risk stratification by the Mayo Molecular Model included; low 0, intermediate-1 25% (n = 4), intermediate-2 37% (n = 6) and high risk 37% (n = 6). There was no difference between DNMT3A mutated and wildtype patients in terms of age and gender distribution, hemoglobin level, leukocyte counts, absolute monocyte counts (AMC), absolute neutrophil counts (ANC), absolute lymphocyte counts (ALC), platelet counts, circulating immature myeloid cells (IMC), peripheral blood (PB) or BM blast content. Concurrent gene mutations were equally distributed with the exception for a higher prevalence of SF3B1 (P = 0.0001) and PTPN11 (P = 0.005) mutations and a lower prevalence of SRSF2 (P = 0.004) mutations in DNMT3A mutated CMML. Five (31%) DNMT3A mutated CMML patients were treated with hypomethylating agents (5-azacitidine- 2, decitabine-3) for a mean of 4 cycles (range, 1–6). Of these, only one achieved a partial response, three had stable disease, while one patient had disease progression. Four (29%) patients underwent leukemic transformation.
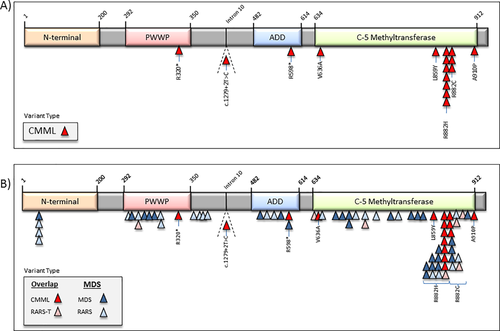
Distribution of DNMT3A molecular variants in patients with myeloid neoplasms. (A) Distribution of DNMT3A molecular variants in 16 patients with DNMT3A mutated chronic myelomonocytic leukemia. (B) Distribution of DNMT3A molecular variants in patients with myelodysplastic syndromes, refractory anemia with ring sideroblasts with or without thrombocytosis and chronic myelomonocytic leukemia. [Color figure can be viewed at wileyonlinelibrary.com]
| Age (years) | Sex | 2016 WHO CMML subtype | Cytogenetic abnormalities | DNMT3A mutations | Concurrent gene mutations | Hypomethylating agent therapy | Outcome | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNMT3A Nucleotide change | DNMT3A Protein change | VAF % | HMA (Y/N) | Number of cycles and best response | Survival in months | ||||||
| 76 | F | CMML-0 | 46,XX[20] | c.2728G>C | A910P | 44 | ZRSR2, SF3B1 | No | 11.9 | Dead | |
| 62 | M | CMML-0 | 46,XY[20] | c.2645G>T | R882C | 51 | TET2 | No | 6.9 | Dead | |
| 87 | M | CMML-0 | 46,XY[20] | c.2644G>A | R882H | 44 | U2AF1 | No | 11.5 | Dead | |
| 57 | M | CMML-2 | 46,XY,del(7)(q22q34) [1]/47,idem,+mar[1] /46,XY[18] | c.2644G>A | R882H | 45 | PTPN11, ASXL1, U2AF1 | Decitabine | 3—SD | 11.1 | Dead |
| 71 | M | CMML-0 | 46,XY[20] | c.2645G>T | R882C | 29 | NRAS, RUNX1, TET2 | No | 10.2 | Dead | |
| 20 | M | CMML-1 | 46,XY[25] | c.1792C>T | R598* | 48 | C-KIT | No | 5.2 | Dead | |
| 66 | F | CMML-0 | 45,XX,-7[19] | c.2644G>A | R882H | 45 | ASXL1, SETPB1 | No | 8.2 | Dead | |
| 60 | M | CMML-0 | 46,XY,del(5)(q13q33),-7,der(11)t(11;?)(q13;?) hsr(11)(q13),der(13) del(13) (q12q14)t(7;13) (q11.2;p13),+r[16] | c.2644G>A | R882H | 51 | TP53 | Decitabine | 1-SD | 0.2 | Dead |
| 34 | M | CMML-1 | 46,XY[20] | c.2644G>A | R882H | 44 | ASXL1, CBL, SETPB1 | No | 50.3 | Dead | |
| 55 | F | CMML-2 | 46,XX[20] | c.2645G>T | R882C | 45 | ASXL1, FLT3-TKD, NPM1 | Decitabine | 5-PR | 6.8 | Dead |
| 67 | F | CMML-0 | 46,XX[20] | c.958C>T | R320* | 59 | SF3B1, TET2 | No | 17.7 | Alive | |
| 73 | F | CMML-0 | 46,XX[18] | c.1907T>C | V636A | 50 | SF3B1, TET2 | No | 24.2 | Alive | |
| 63 | M | CMML-0 | 46,XY[20] | c.2644G>A | R882H | 46 | ASXL1, RUNX1, SRSF2, TET2 | Azacitidine | 6-SD | 28 | Alive |
| 72 | F | CMML-0 | 45,XX,-7[8]/46,XX[12] | c.1279 + 2T>C | 44 | ASXL1, NRAS, RUNX1, TET2 | No | 9 | Alive | ||
| 80 | M | CMML-0 | 46,XY [30] | c.2644G>A | R882H | 52 | JAK2, SF3B1,TET2 | Azacitidine | 4- P | 26 | Dead |
| 63 | F | CMML-1 | 45,XX,add(1)(p32),psu dic(5;6)(q11.2;q13),-7,der(12;13)(q10;q10), +2mar[20] | c.2576L>T | L859Y | 43 | PTPN11, TET2, TP53 | No | 3.6 | Alive | |
- CMML, chronic myelomonocytic leukemia; M, male, F, female; VAF, variant allele frequency burden; HMA, hypomethylating agents; SD, stable disease, PR, partial response; P, progression.
Impact on OS and leukemia-free survival
The median survival for the entire cohort (n = 261) was 24 months. In univariate analysis, survival was shorter in DNMT3A mutated (median 8 months) versus wildtype (median 27 months) patients (P = 0.0007; HR 2.9, 95% CI 1.5–5.7; Fig. 2A). Other variables of significance, in univariate analysis, included lower hemoglobin (P = 0.002), higher leukocyte count (P = 0.0009), higher AMC (P = 0.0012), PB blast% (P = 0.001), circulating IMC (P = 0.01), BM blast% (P = 0.045), abnormal karyotype (P = 0.02), and ASXL1 (P = 0.01) mutations. Survival was also adversely affected by the presence of either (n = 133) or both (n = 3) ASXL1/DNMT3A mutations (0 = 0.007, Fig. 2B). In multivariable analysis excluding ASXL1 and DNMT3A mutations, hemoglobin (P = 0.03), IMC (P = 0.013) and AMC (P = 0.02) retained significance. When ASXL1 mutations were added to the multivariable analysis, ASXL1 (P = 0.01) mutations, AMC (P = 0.012) and IMC (P = 0.03) retained significance. Similarly, when only DNMT3A mutations were added to the multivariable analysis, DNMT3A (P = 0.003) mutations, IMC (P = 0.01), and AMC (P = 0.02) retained significance. When both DNMT3A and ASXL1 mutations were added to the multivariable analysis, only DNMT3A (P < 0.0001) and ASXL1 (P = 0.004) mutations remained significant. DNMT3A mutations predicted shortened survival, independent of the ASXL1 inclusive GFM model (P < 0.0001) and the Mayo Molecular Model (P = 0.002).
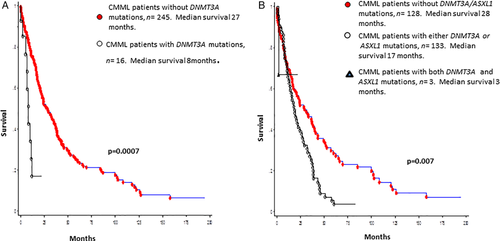
(A) Survival data for 261 patients with WHO defined chronic myelomonocytic leukemia stratified by their DNMT3A mutational status. (B) Survival data for 261 patients with WHO defined chronic myelomonocytic leukemia stratified by their DNMT3A and ASXL1 mutational status. [Color figure can be viewed at wileyonlinelibrary.com]
In comparison to their wildtype counterparts, LFS was significantly shorter in DNMT3A mutated patients (16 versus 4 months, P = 0.02) (Fig. 3). On a univariate LFS analysis that included age, gender, hemoglobin, leukocyte count, AMC, ANC, circulating IMC, platelet counts, PB and BM blast counts, 28 myeloid-relevant gene mutations, karyotype and WHO CMML morphological subtypes; only low hemoglobin level (P = 0.01) and DNMT3A mutations (P = 0.0018), predicted for shortened LFS. In a multivariable model that included low hemoglobin [P = 0.003, HR 1.3, 95% CI 1.1–1.6] and DNMT3A mutations [P = 0.003, HR 1.52, 95% CI 1.3–1.5], both variables retained independent prognostic significance.
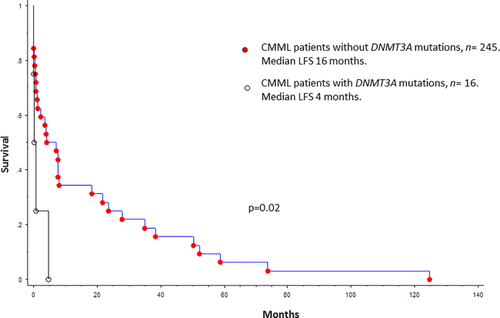
Leukemia-free survival data for 261 patients with WHO defined chronic myelomonocytic leukemia stratified by their DNMT3A mutational status. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
DNA methylation is mediated by a family of DNA methyltransferase enzymes that include DNMT1, DNMT3A, DNMT3B, and DNMTL 8. Whereas DNMT1 maintains pre-existing DNA methylation patterns, DNMT3A/3B participate in de novo DNA methylation 8. DNMT3L, lacks a catalytic domain, functions as an accessory protein to DNMT3A and is mainly seen during embryonic development and during genomic imprinting 8. While DNMT3A mutations are common in hematological malignancies, they have also been described as a component of age related clonal hematopoiesis 18. In a large population-based study (n = 17,182), somatic variations involving DNMT3A (variants = 403), TET2 (variants = 72) and ASXL1 (variants = 62) were seen in ∼11% of the population >80 years of age, and in comparison to patients without clonal hematopoiesis, were associated with an increased risk of hematological malignancies (HR—11.1) and all-cause mortality (HR-3.7) 18.
DNMT3A mutations are frequent in AML, occur in ∼22% of cases (∼35% in cytogenetically normal AML), with some, but not all studies demonstrating an adverse impact on survival 19, 20. Similarly, in MDS, DNMT3A mutations occur in ∼13%, tend to cluster with SF3B1 mutations (negative association with SRSF2 mutations), with an inconsistent impact on survival.21, 22 In MPN, DNMT3A mutations are seen in ∼10%, are more common in myelofibrosis (∼6%) and blast transformed MPN (∼15%), and once again have an unclear prognostic impact 11. In CMML, DNMT3A mutations are seen in ∼2–5% of cases, most commonly involve the Arg882 hot spot and are more common in CMML-2 and blast transformed CMML (∼26%) 6, 23. In our study, DNMT3A mutations were observed in 6% of patients, were predominantly missense (81%), mainly (81%) affected the catalytic methyltransferase domain, with 63% (n = 11) involving the Arg882 mutational hot spot. The Arg882 substitution results in a hypomorphic protein that acts in a dominant negative manner, inhibiting the methyltransferase activity of the remaining wildtype DNMT3A 24. This mutation correlates with global hypomethylation at CpG islands, shores and promoters; although some promoter hypermethylation has also been reported 8, 25. Mutations affecting the ADD and the PWWP domains were less frequent (∼6% each). The ADD domain interacts with important regulatory elements including H(histone)3K(lysine)4me(methyl)0, HDAC1 (histone deacetylase 1), SETDB1 (histone-lysine N-methyltransferase), H4R(arginine)3me2, and EZH2 (enhancer of zeste homolog 2); while the PWWP domain interacts with DNA, heterochromatin, G9A (histone methyltransferase), H3K36me3 and H3K9me3, respectively 8. Unlike in other myeloid neoplasms, there were no mutations affecting the N-terminal domain. The N-terminal domain interacts with DNA, proteins, Np95 (histone binding protein with ubiquitin ligase activity) and K44me 8.
Concurrent gene mutations were ubiquitous (100%), and as in MDS, DNMT3A mutations clustered with SF3B1 mutations (P = 0.0001) and had a negative association with SRSF2 mutations (P = 0.004). All DNMT3A/SF3B1 comutated CMML patients had bone marrow ring sideroblasts (RS). While SRSF2 mutations are the most frequent spliceosome component mutation in CMML (∼45%) 26, SF3B1 mutations are common in MDS with RS (∼80%), and can be seen in CMML patients with RS (<10%) 26, 27. The clustering of DNMT3A mutations with SF3B1 mutations in CMML raises the question as to whether this group of patients has a unique molecular signature. We also demonstrate a significant correlation between DNTM3A and PTPN11 mutations (P = 0.005). This association has been reported in de novo AML (n = 506), where 10% of DNMT3A mutated patients had concurrent PTPN11 mutations (P = 0.007) 28. PTPN11 codes for a protein tyrosine phosphatase that plays an important role in signal transduction (including the RAS pathway) and the significance of this association remains to be elucidated.
Given the low frequency of DNMT3A mutations in CMML, the prognostic impact of these mutations have not been well defined. Jankowska et al. identified DNMT3A mutations in 4% of their cohort (n = 72), and on univariable (P = 0.04), but not multivariate analysis, found them to negatively impact survival 23. In a large GFM study (n = 312), DNMT3A mutations were seen in 2% of patients, and given their low frequency were not included in further survival analyses 6. In the recent CPSS-Mol study, DNTM3A mutations were seen in 4% and did not impact survival 7. In our prior Mayo Clinic study (n = 175), DNMT3A mutations were seen in 5% (n = 9), and on univariable (P = 0.02), but not multivariate analysis, were associated with shortened survival 3. In the current study which included a larger number of informative cases (n = 261), we demonstrate the negative prognostic impact of DNMT3A mutations on both overall survival and leukemia-free survival. This finding was significant in a multivariate analysis that included other CMML relevant prognostic variables, including nonsense and frameshift ASXL1 mutations. In addition, DNMT3A mutations predicted shortened survival independent of the ASXL1 inclusive GFM model and the Mayo Molecular Model. This finding warrants inclusion of DNMT3A mutations in molecularly integrated CMML prognostic models.
Acknowledgments
Current publication is supported in part by grants from the “The Henry J. Predolin Foundation for Research in Leukemia, Mayo Clinic, Rochester, MN, USA”.
This publication was supported by CTSA Grant Number KL2 TR000136 from the National Center for Advancing Translational Science (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.




