3D bioprinted organ-on-chips
Abstract
Organ-on-a-chip (OOC) platforms recapitulate human in vivo-like conditions more realistically compared to many animal models and conventional two-dimensional cell cultures. OOC setups benefit from continuous perfusion of cell cultures through microfluidic channels, which promotes cell viability and activities. Moreover, microfluidic chips allow the integration of biosensors for real-time monitoring and analysis of cell interactions and responses to administered drugs. Three-dimensional (3D) bioprinting enables the fabrication of multicell OOC platforms with sophisticated 3D structures that more closely mimic human tissues. 3D-bioprinted OOC platforms are promising tools for understanding the functions of organs, disruptive influences of diseases on organ functionality, and screening the efficacy as well as toxicity of drugs on organs. Here, common 3D bioprinting techniques, advantages, and limitations of each method are reviewed. Additionally, recent advances, applications, and potentials of 3D-bioprinted OOC platforms for emulating various human organs are presented. Last, current challenges and future perspectives of OOC platforms are discussed.
1 INTRODUCTION
One of the great constraints toward therapeutical practices is that most of the available reported approaches for medical trials have been performed in animal models (e.g., mice) or static two-dimensional (2D) cell-culture phantoms. However, animal testing faces challenges due to its low throughput, lack of cost-effectiveness (feeding, housing, and animal care costs), ethical concerns, differences in target homology, and dissimilarities in physiology, resulting in therapeutic strategies with affirmative effects in animal models while oftentimes not achieving the same outcome in humans.[1-5] For instance, the majority of mouse models studied for the treatment of coronavirus disease 2019 (COVID-19), a worldwide pandemic, failed to illuminate all facets in humans, particularly unfamiliar futures such as pulmonary vascular disease and hyperinflammatory syndromes.[6] On the other hand, despite their simplicity in culture, effectiveness, and widespread use in research, 2D cell cultures face challenges in controlling cell culture structure, in mimicking cell–cell interactions and biochemical signals, as well as in modeling mechanical microenvironments of real tissues and cell polarity, since the 2D microenvironment has limitations in representing the physiological conditions of the human body in most cases.[2, 7-11] Recent studies have attempted to address a number of the above challenges such as shape control using microwells[12] as well as micropillars,[13] and cell polarization effects using a sandwich culture method[11, 14-16] (i.e., adding an extracellular matrix (ECM) layer over cells to eliminate apical-basal polarity).[11] Nevertheless, 2D cultures are generally considered to be limited in sufficiently mimicking in vivo conditions.
Compared with 2D cultures, 3D cell cultures more realistically recapitulate the complex structures of human tissues by allowing cells to cluster, migrate, polarize, and expand, while regenerating in vivo-like cell signaling pathways, functions, and drug responses by simulating the hindering effect of ECM and/or outer layer of cells on the diffusion of drug molecules in vivo.[17-21] However, conventional 3D cultures cannot replicate the dynamic microenvironment of the human body, such as biofluid flow (e.g., blood and lymphatic fluid for waste removal, nutrient supply, and drug transport), spatiotemporal oxygen distribution, and mechanical stresses experienced by cells during respiration and heartbeat.[18, 22]. An alternative perspective to overcome these challenges can be demonstrated by the use of organ-on-a-chip (OOC) platforms based on human cells.[23] This eccentric approach, with its innovative ability to provide insights into real-world human organ functionality and disease pathophysiology, along with a more accurate prediction of efficacy and potential side effects of a new drug or therapeutic practice, offers valuable integration into translational science and regenerative medicine.[11] The use of cell culture for disease studies in vitro dates back to 1991, when one of the first papers in this context was published discussing organized cell culture for the construction of ventricular myocardium, which allowed the biophysical elucidation of conduction block in the heart.[24, 25] In 2004, the concept of mimicking human physiology on a microfluidic chip was introduced to study the systemic interaction between liver and lung on a silicon chip.[26, 27] The term “organ-on-chip” was first coined in 2010 for a microfluidic chip used to study the human lung at the organ level.[28]
OOC systems are convenient, versatile means of mimicking the functions of various organs of the human body with the ability to be seeded with human cells to create patient-specific, multicellular setups for conducting personalized medicine research and an environment for studying realistic organ interactions with proposed therapeutic approaches.[29-34] The main advantages offered by microchannels, chambers, valves, and pumps, for cell culture, may include perfusability and possible gas permeability (which increase cell viability and metabolic rate), transparency (which enables microscopic imaging),[35, 36] integrability with sensors (which allows real-time screening of culture, biomarkers, and responses to stimuli),[37, 38] gradient generation as a result of laminar flow in microchannels (which enables the study of differentiation and directed cell migration), porous membranes (modeling tissue barrier functions, transcellular transport, secretion, and absorption), cost-efficiency (lower volume of expensive samples/reagents due to microscale channels), sophisticated structures (wide range of manufacturable geometries on microfluidic chips), mimicking of dynamic in vivo conditions (emulating cyclic mechanical stress and strain experienced by cells during peristalsis, respiration, and cardiovascular cycling), and/or single-cell analysis.[5, 10, 29] Conventional OOC fabrication approaches (e.g., soft lithography, microcontact printing, and replica molding[39, 40]) usually require cleanrooms, a high level of microfabrication expertise,[41, 42] a secondary cell-seeding step (resulting in intense protein absorption), and have problems implementing cell–cell and cell–ECM interactions to emulate spatial heterogeneity.[8]
Among the various approaches for fabrication of OOCs, 3D (bio)printing has the potential to provide a fabrication method for simultaneous/consecutive generation of sophisticated structures of ECMs and cells (e.g., patient-derived cells) with a fast turn-around time as well as great freedom to modify the chip design.[2, 43] It is expected that the 3D bioprinting market for the medical and healthcare sectors will increase at a compound annual growth rate (CAGR) of 14.5% from 2021 to 2028, reaching revenue of up to $4.4 billion in 2028.[44] Bioprinting is a branch of the well-known 3D printing process in which a computer-aided design (CAD) is processed, typically in a layer-by-layer manner, to complete the structure of the product using solidifiable biomaterial, including but not limited to ion-crosslinkable, temperature-sensitive, and photopolymer bioinks.[45, 46] Valve-based[47] and inkjet-based,[48-50] acoustic,[51, 52] microextrusion,[53-55] and light-enabled[56-58] bioprinting are commonly used technologies.[59]
Over the years, 3D printing has successfully contributed to the fabrication of medical devices,[60-64] sensors,[65] tissue scaffolds,[66] and microfluidic chips[67-69] for sensing, gradient generation, chemical mixing, tissue engineering, and OOC applications.[8] Although cells are not usually present in 3D printing, the use of cytocompatible biomaterials enables 3D bioprinters to directly print with cells; alternatively, cells can be bioprinted without additional biomaterials.[70] Integrating the advantages of microfluidic chips (e.g., gas permeability, perfusion, and single-cell analysis) with 3D bioprinting can lead to automated bioprinting of reproducible, precisely positioned, and perfused multicell cultures, with customized structure/features (e.g., pore size and morphology), for physiological studies as well as drug analysis at the organ level,[71] such as kidney-,[72, 73] heart/vasculature-,[74-76] liver-,[77] brain/blood–brain barrier (BBB)-,[78-81] bone/cartilage-,[82, 83] cancer/tumor-,[84, 85] placenta-,[86] gut-,[87] and lung-[88] on-chips.
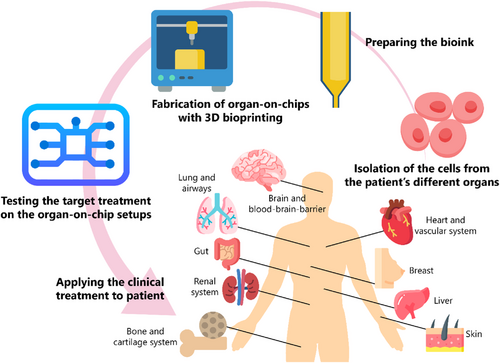
Here, we review recent advances in 3D-bioprinted OOCs. First, the different bioprinting technologies are presented, as well as the advantages and disadvantages of each process. In addition, bioinks and cell sources used in these methods are discussed. Subsequently, examples of 3D-bioprinted OOCs are reviewed, as shown in Figure 1, including applications of OOCs in the cardiovascular system, brain and BBB, lung and respiratory system, liver, gut, renal system, breast, bone and cartilage system, and skin studies, highlighting the design features, advantages, and limitations of the cases studied. Finally, challenges and future research areas are presented.
2 FABRICATION
2.1 Bioprinting
The conventional fabrication techniques of OOCs include photolithography, soft lithography,[89-91] replica molding,[92] capillary molding,[89] microcontact printing,[92] microtransfer molding,[89] and injection molding.[93-95] One of the limitations of the above methods is the limited fabrication capability for forming the complex structures of organs and tissues.[96, 97] On the other hand, these methods usually require multistep production protocols. In particular, lithographic techniques need to be carried out through several lithographic processes and masks.[29, 96, 98] This leads to experiments that are time consuming and expensive. In addition, the traditional methods require a secondary organization for the cell seeding process, which oftentimes drives up the overall cost, as well as poor selectivity of different cell types.
The adoption of 3D printing and 3D bioprinting in medical and biomedical applications has resulted in cost efficiency, rapid turnaround times, and a wide range of materials.[99-103] Moreover, prototyping of OOCs with 3D bioprinting requires minimal microfabrication skills and enables simultaneous/consecutive (bio)printing of polymers, hydrogels, and multiple cell types to produce customized, reproducible, perfusable, and complex patient-specific 3D biomimetic tissue constructs with high precision in the placement of cells,[8, 29, 43, 96] which is hardly achievable with the conventional techniques. 3D bioprinting techniques can be primarily divided into two categories (Figure 2): (i) nozzle-based (e.g., inkjet-based (droplet-based) and extrusion-based bioprinting) and (ii) light-enabled bioprinting (e.g., stereolithography apparatus (SLA)-/digital light processing (DLP)-based bioprinting, two-photon polymerization (TPP)-based bioprinting, laser-assisted bioprinting, and computed axial lithography).[104-106] A summary of the advantages, limitations, and important properties of the commonly used 3D bioprinting methods is shown in Table 1.

| Method | Extrusion-based bioprinting | Inkjet bioprinting | Stereolithography-based bioprinting (SLA, DLP) | Multiphoton-based bioprinting | Laser-based bioprinting |
|---|---|---|---|---|---|
| Working principle |
Pneumatic, mechanical, microfluidics-assisted |
Thermal, piezoelectric, electrostatic |
Point-by-point exposure (SLA), layer-by-layer exposure (DLP) | Point-by-point exposure by a light beam | Laser-induced |
| Printable viscosity | 30 to 6 × 107 mPa · s[104] | 3–12 mPa · s[104] | 1-1,000 mPa · s[333] | – | 1–300 mPa · s[71, 334] |
| Cell density | High[334] | Low[334] | Medium[334] | Medium | Medium[334] |
| Print speed | Medium[72, 335] | High[335] | Medium to high[71] | High | High |
| Resolution | >100 μm[118] | >10 μm[141] | >6–10 μm[71] | >100 nm[336] | >50–100 μm[337] |
| Cell viability | >90%[115] | >85%[338] | ∼80%[144] | >90%[152] | >90%[58, 154] |
| Advantages | Affordability, large deposition rate, bioprintability of high viscosity bioinks with high cell concentration, simultaneous bioprinting, scalability[72, 118] | Affordability, good resolution, fast bioprinting, the ability to generate a cell concentration gradient[71, 334] |
High resolution, oftentimes high speed, no clogging problem, bioinks with high cell concentration can be bioprinted[71, 334] | High resolution, multidirectional bioprinting, good viability[336] | High resolution (single cell per droplet), high cell viability, no clogging, ability to bioprint low-viscosity bioinks with high cell density[334, 337] |
| Limitations | High shear stress, nozzle clogging, low resolution, low structural fidelity, limited material choice (only materials with shear-thinning property)[72, 118] |
High-viscosity bioink cannot be bioprinted, limited vertical bioprinting, clogging, shear stress, additional cross-linking step is required for low viscosity bioinks[71, 334] |
Limited choice of photosensitive material, Possibility of cell lysis or DNA damage, expensive equipment[71, 334] |
High cost, limited biomaterial choice, near-infrared transparency requirement, low bioprinting speed[336] | High cost, cytotoxicity induced by metallic nanoparticles, photonic cell damage, complex control of laser pulses[334, 337] |
2.1.1 Nozzle-based methods
The working principle of nozzle-based bioprinting is based on the ejection of bioink through a nozzle by applying a force field.[107] As one of the most studied and available types of bioprinting, nozzle-based bioprinting is a low-cost method with a moderate resolution and longer processing times compared with optical methods.[108, 109] In nozzle-based bioprinting, the amount of shear stress experienced by the bioink, along with temperature, are the major factors that threaten cell viability and limit the applicability of nozzle-based bioprinting for cell/tissue bioprinting.[110, 111] Nozzle-based bioprinting methods can be classified into extrusion-based and droplet-based methods. Droplet-based methods can be further divided into microvalve-based bioprinting, acoustic droplet bioprinting, inkjet bioprinting (continuous inkjet (CIJ), drop-on-demand (DoD) (thermal, piezoelectric, and electrostatic)), and electrohydrodynamic jetting.[71]
2.1.1.1 Extrusion-based bioprinting
In extrusion-based 3D bioprinting, the bioink is typically placed in a syringe-like tool that uses a controlled force—pneumatic pressure or mechanical force generated by a piston or screw—to propel the bioink through the nozzle[112-114] (Figure 2A). This bioprinting technique can deposit highly viscous bioinks with high cell concentrations at a large deposition rate. Moreover, multinozzle extrusion bioprinters enable the simultaneous deposition of heterogeneous structures. However, the resolution of this method is usually limited. In extrusion bioprinting, the main factor affecting cell viability is the shear stress acting on the bioink, which depends on the viscosity of the bioink, nozzle dimension, and printing pressure.[115] Although printing resolution can be increased by smaller nozzles, a decrease in nozzle diameter results in higher shear stress (i.e., lower cell viability) and nozzle clogging. Besides, the use of highly viscous bioinks or a high flow rate of bioink in the nozzle can also increase shear stress. Another limitation is that the bioink used in extrusion-based bioprinters should ideally have a shear-thinning property, which limits the choice of bioink.[109, 116] Furthermore, depending on the used bioink, the constructs bioprinted by extrusion-based methods may suffer from low structural fidelity and undesirable deformation in long-term cultures due to the junctional seams and voids created by line-by-line deposition of the cylindrical filaments.[117]
A multihead extrusion bioprinting system, with a resolution of 50 μm, was developed for bioprinting human tissues using cell-loaded hydrogels through a 300-μm Teflon nozzle at a pressure of 50–80 kPa, yielding >90% cell viability for bioprinting bone, cartilage, and muscle tissues.[118] In another experiment, using four autonomously addressable printheads (with diameters of 100–410 μm), a perfusable tissue (with >95% cell viability) was bioprinted on a chip at printing speeds ranging from 1 mm/s to 5 cm/s respective to air pressures ranging from 10 to 140 psi.[119] To study the effects of bioink viscosity on bioprinted tissue, a bioprinter with a painting needle was designed to bioprint cardiac tissue on a chip with ∼92% cell viability using bioinks with viscosities ranging from 1 to 1 × 105 mPa · s. The setup was fast (extruding up to once every 0.1 s) with precise control over the volume of extruded bioink (several hundred nl to several pl). By controlling the number of ladened cells by altering the time and diameter of the painting needle, a high-density (3.5 × 108 cells/cm3) 3D cardiac tissue was bioprinted.[120] Integration of microfluidic printheads with extrusion-based bioprinters can miniaturize shear stress and promote cell viability.[111] The microfluidic printhead enables simultaneous extrusion of bioink and crosslinker, while sheathing cells by crosslinker on either side, creating a shear stress-protective layer between the nozzle wall and cells during the extrusion process.[111] In another study, neural constructs were coaxially bioprinted by separate micropumping of crosslinker and bioink. Using 3–5 μL of bioink, cell layers of 100 μL were bioprinted at a speed of 240 mm/min.[121] Extrusion-based bioprinting represents a convenient, intensively explored rapid prototyping platform with reasonable resolutions and costs (in the abovementioned studies, although cells were not necessarily bioprinted on a chip, the achieved bioprinting resolution demonstrates the potency of these methods for the fabrication of 3D-bioprinted OOC platforms in the future).
2.1.1.2 Inkjet bioprinting
Inkjet bioprinting has been largely adapted from commercial inkjet printers, making it available in affordable price ranges. The bioink is filled into a chamber that has a nozzle (inkjet heads) and an actuation mechanism, which can be a thermal,[122] piezoelectric,[123] or electrostatic,[124] mechanism (Figure 2B). Two main categories of inkjet bioprinters are CJI and DoD. Although both deposit droplets of bioink on the surface, CIJ creates a stream of bioink droplets by forming Rayleigh-Plateau instability,[125] which limits the precise control of droplet position.[126] On the other hand, DoD inkjet bioprinters produce droplets only when the discharge signal is present, resulting in better resolution and effective bioink utilization, making DoD a more suitable method for OOC bioprinting.[50] Depending on the actuation mechanism, the critical parameters affecting cell viability are temperature, electric field, and shear stress.[49, 107, 127, 128] Affordability, higher printing speeds (up to 10,000 droplets per second), and higher resolutions (∼50 μm) compared with extrusion-based bioprinting are the main advantages of inkjet bioprinters.[129] A weakness of inkjet bioprinting is that bioinks with high viscosities cannot be bioprinted properly because they can cause clogging at the outlet nozzle. This leads to a lower number of cells delivered per unit time, as a result of using low-viscosity bioinks with lower cell content.[120] Furthermore, since inkjet bioprinters can only operate with low-viscosity bioinks (∼3–12 mPa · s), an additional crosslinking step is almost always required after bioprinting to achieve a stable structure.[71, 130]
A DoD inkjet bioprinter (with a 300-μm microvalve and 450-μs valve opening time) was used to bioprint neural progenitor cells and spheroid breast cancer cells through a flat-tip 27 G needle at varying pressures from 0.25 to 1.5 bar. Both tissue-on-chips retained their viability for up to 14 days.[131] Another DoD bioprinter with two 300-μm microvalves and a 150-μm electromagnetic microvalve, with an opening time of 450 μs, was used to bioprint bioink, channel wall material, and crosslinker under 0.5 bar of pressure to create a perfusable vessel model with >83% cell viability.[132] Another on-demand inkjet bioprinter was developed to electrohydrodynamically bioprint tumor cell-laden hydrogel array (>90% cell viability) onto an array chip for drug screening through a 32 G nozzle at 2.7 kV and a flow rate of 10 μL/min.[17] To take advantage of both extrusion-based methods, a hybrid method was proposed that uses extrusion-based printing to print a collagen-based transwell construct and inkjet bioprinting to uniformly bioprint keratinocytes onto this structure, successfully forming a skin that recapitulates in vivo biological properties.[133]
2.1.2 Light-enabled methods
SLA-based bioprinting methods are mainly adapted from lithographic methods used in semiconductor fabrication. In this bioprinting method, a photosensitive bioink is exposed to the light of a specific wavelength to polymerize (in the case of negative photoresist) or depolymerize (in the case of positive photoresist) the exposed areas.[134, 135] Although nozzle-based processes typically employ physical (e.g., heat) and chemical crosslinking, SLA uses photocrosslinking which has better spatial and temporal control with minimal heat generation and faster room-temperature progression during the polymerization process.[136] SLA printers cure photoresists in a point-by-point manner, which prolongs the printing process. Although the use of physical photomasks enables the exposure of a layer at once to increase the printing speed, preparing different photomasks is cumbersome. SLA was made more suitable for bioprinting by the adoption of DLP or liquid crystal display SLA (LCD-SLA) to eliminate the use of physical photomasks[137, 138] (Figure 2C). These methods cure the desired pattern by either using an array of several thousand independently controllable digital micromirror devices (DMDs) or using the light from an array of LEDs masked by LCD to form a 2D image of slices to perform a layer-in-once exposure, ultimately improving resolution and printing speed.[138-140] An important factor for cell viability and print resolution in SLA printing is the wavelength, which is mostly in the ultraviolet (UV) range.[141] The DLP method oftentimes uses visible light as a light source, reducing the risk of cell damage.[142] Moreover, since SLA/DLP is a nozzle-free method, bioinks can be used without clogging problems. The main challenges in using SLA/DLP are the limited choice of photopolymerizable bioinks, the possibility of deoxyribonucleic acid (DNA) damage as well as cell lysis as a consequence of UV exposure, and the expensive equipment.[134] Nonetheless, the use of visible light for bioink curation (using visible-light photoinitiators) could partially address problems that arise from UV exposure.[143]
DLP-based bioprinting was used to bioprint bone-mimetic structures in vitro.[142] Using a DMD 3D bioprinter, human hepatocellular carcinoma (HepG2) cells were bioprinted with viability greater than 80%.[144] Moreover, human induced pluripotent stem cell (hiPSC)-derived hepatic cells were bioprinted in gelatin methacryloyl (GelMA) using a 365-nm light and showed 76% viability 2 h after printing. Oxygen distribution in the hydrogel, as a factor of the thickness of the structure, was controlled by tuning the exposure time/intensity with a motion controller.[145] A microfluidic chip was integrated with a DMD bioprinter (using 365 nm, 500-mW/cm2 light) to achieve printing resolutions of 100 μm in z- and 10 μm in x-, y-directions. The microfluidic chip with a PDMS chamber and four inlets allowed sequential injection of different bioinks to produce a multimaterial bioprinting platform.[84] Additionally, an DMD system was used for direct 3D bioprinting of cell-laden constructs in microfluidic architectures (10T1/2 cells in GelMA).[146] In another light-enabled study, a commercial projector was used as a light source to bioprint lung adenocarcinoma cells with a resolution of 38 μm at a wavelength of 405 nm. The manual saline rinsing step and automated material selection process resulted in heterogeneous structures without undesired mixing (in the abovementioned studies, although cells were not bioprinted on a chip, the achieved bioprinting resolution demonstrates the potency of these methods for the fabrication of 3D-bioprinted OOC platforms in the future).[117]
Another approach to light-induced polymerization is TPP/multiphoton polymerization (TPP/MPP), also known as direct laser writing (Figure 2D).[60] Biocompatible, high resolution (∼100 nm), and selective consolidation of photosensitive materials are possible by exposure of focused (point by point) low-energy femtosecond (fs) laser pulses (near-infrared (NIR) light) with the ability to direct laser focal point in the desired direction.[60, 147-149] However, since polymerization is point-by-point like in SLA, the printing speed is generally lower than DLP. The technique has been used to produce scaffolds in biocompatible hydrogels with a focus spot size of 5 μm using 0.1–10 nJ per pulse, infrared pulses (100-fs pulses of λ = 810 nm) at 80 MHz.[150] The transmissivity of most cells to NIR light allows MPP to form 3D structures within cell-ladened hydrogels and even within tissue/body, with minimal cell damage.[147, 151] When choosing MPP as a bioprinting method, it should be considered that despite the high resolution, the choice of biocompatible materials for MPP is limited.[152]
2.1.3 Laser-based 3D bioprinting
Laser-based bioprinting techniques use CAD to pattern cell-laden bioinks using laser energy.[58] The main components of a laser-based bioprinter are a laser source (continuous or pulsed), a laser-transparent printing ribbon coated with a layer of cell-laden bioink, and a substrate slide placed on a movable plate (Figure 2E). Based on the type of these components, laser-based methods can be divided into laser-guided direct-writing, matrix-assisted pulsed laser evaporation direct writing, biological laser processing, absorbing film-assisted laser-induced forward transfer, and laser-induced forward transfer (LIFT). LIFT is one of the commonly used bioprinters for tissue fabrication.[56, 153] Although SLA, TPP/MPP, and LIFT methods use a laser beam for printing, SLA and TPP/MPP use the laser to polymerize the photosensitive bioink. However, in the LIFT process, a laser beam (a pulsed NIR laser beam) is focused on a glass substrate coated with an absorbent layer (usually Au), and the energy generated at this point produces a microdroplet from the cell-laden coated bioink layer.[154, 155] Notable advantages of laser-based bioprinters include high resolution (with the ability to bioprint single cell per droplet), high cell viability after bioprinting (∼95%), no clogging, the ability to bioprint low-viscosity bioinks (1–300 mPa · s), and the capability to bioprint cell suspensions with high cell density (up to 108 cells per mL).[71, 136] On the other hand, the risk of photonic cell damage (as a result of laser radiation), the possibility of cytotoxicity induced by metallic nanoparticles[156] (as a result of using metals as laser energy-absorbing layer), the complexity of controlling laser pulses, the difficulty of manufacturing the ribbons, and the high cost of laser systems are the main limitations of laser-based bioprinting.[71] Initial studies reported 98 and 90% of cell viability for skin cells and human mesenchymal stem cells (hMSCs), respectively, after bioprinting using the LIFT method.[157] Using a laser-assisted 3D bioprinter, exocrine pancreatic spheroids were bioprinted with a wavelength of 1064 nm, a pulse of 30 ns, and a repetition rate of 1–100 kHz, reporting a survival rate of >90%, 24 h after bioprinting.[154]
2.2 Bioink
Solidifiable materials, including ion-crosslinkable hydrogels, temperature-sensitive polymers, and photopolymer bioinks are commonly used materials for 3D bioprinting.[2, 158] The bioink can be derived from natural sources, such as alginate (also termed algin or alginic acid), carrageenan (also termed carrageenin), gellan gum, agar (also termed agarose), collagen, fibrin, gelatin, silk, fibrinogen, chitosan, methyl cellulose (derived from cellulose), and hyaluronan,[159-162] or it can be derived synthetically, such as poloxamer (also termed Pluronic), Matrigel, poly(caprolactone) (PCL), poly(ethylene glycol) (PEG), GelMA, poly(vinyl alcohol) (PVA), poly(lactic acid) (PLA), and poly(lactic-co-glycolic acid) (PLGA).[161, 162]
Alginate (a natural polymer extracted from brown seaweeds) is a cost-efficient option for bioink preparation with no toxicity, and good printability, yet with low cellular adhesion and slow degradation.[163] Although carrageenan (a natural polysaccharide extracted from red seaweeds) is another bioink with fine biocompatibility, bioactive, mechanical, and rheological properties, its rather high toxicity is challenging.[164-166] Gellan gum (an anionic microbial polysaccharide) is a cost-efficient biomaterial that offers shear-thinning properties, and high gelling efficiency, while suffering from limited printing fidelity.[167] Agar (a polysaccharide isolated from sea kelp) is another bioink candidate that, despite having high mechanical strength and cost-efficiency, has limited cell adhesion.[168] Collagen (one of the well-known body proteins) also can be used as bioink, offering good cell adhesion as well as growth, and facing challenges of low viscosity and weak mechanical properties.[161] However, although gelatin has the same pitfalls as collagen, gelatin as a collagen-degradation product, offers nonimmunogenicity while maintaining the cell-friendly binding regions.[161] Silk (a natural protein fiber) possesses biodegradability, nontoxic degradation residues, and high cellular viability, whereas it suffers from limited cell growth and function.[61, 161, 169] Matrigel (solubilized basement membrane matrix secreted by Engelbreth-Holm-Swarm mouse sarcoma cells), although being expensive and unsuitable for translation into clinical setups, promotes cell growth and differentiation.[170] PVA is a biocompatible, water-soluble polymer with suitable hydrophilicity and toughness for bioink preparation that has limitations in the adhesion of cells.[161, 171] PEG (one of the FDA-approved materials for medical applications) is a biocompatible and easily modifiable polymer, well known for usage as sacrificial bioink, with limitations in mechanical strength and cell adhesion.[161, 169] Furthermore, GelMA is a biocompatible and biodegradable option for the preparation of bioinks, where a crosslinking process with UV light is needed that can adversely affect cell viability, although visible-light options are becoming more broadly adopted.[161, 172]
One of the most important factors in selecting one of these materials for the preparation of the desired OOC is the crosslinking mechanism, which should be chosen to be harmless to the cells to be loaded into the bioink, based on the fabrication method (photosensitive bioinks for light-induced methods and bioinks with shear-thinning ability for nozzle-based approaches). Cell interaction is another decisive factor for material selection. For example, bioinks made of collagen and hyaluronic acid are superior to alginate and silk fibroin, as they provide better cell interactions.[159] For cancer modeling on a chip, the GelMA hydrogel is a suitable candidate as it promotes cell functions (e.g., tumor cell metastasis and invasiveness), rapid crosslinking, and biocompatibility.[17] Biodegradability, on the other hand, may be important for certain applications. In these cases, it is recommended to use materials with a higher degradation rate compared with other bioinks, such as hyaluronic acid. In addition, silk fibroin usually shows better mechanical properties compared with the other bioinks.[159] Bioinks should be able to maintain their mechanical integrity over a long incubation period under culture conditions (e.g., pressure, temperature, and humidity).[173] An important role of bioinks is to encapsulate the cells to protect them during the bioprinting process, particularly in nozzle-based processes. Hybrid bioinks are promising innovations to improve the mechanical properties of available bioinks.[174, 175] For instance, pure alginate, although readily processable, biocompatible, and widely available, has poor pattern fidelity and printability. However, the combination of alginate with cellulose nanocrystals (whisker/rod-shaped nanoparticles extracted from the crystalline regions of cellulose fibers and characterized by renewability, high mechanical strength, low density, and cytotoxicity) results in a reinforced bioink with better cell protection as well as shear-thinning properties that can be bioprinted through a 100-μm nozzle without clogging.[176] In addition, composite formulas incorporating biologically derived components and living cells, called “living materials”, have recently gained attention for the specific characteristics that they offer, such as autonomic iridescence capability,[177] especially for the preparation of bioinks for 3D bioprinting processes.[178] Powered by various branches of sciences, such as microfluidics, genetics, and cell coating, living materials enable several applications in living structures and organ models.[178, 179]
2.3 Cell source
The key element of 3D bioprinting is the bioink, which requires to meet some necessities, such as appropriate rheological, biocompatibility, and biological properties, structural cell growth support, and suitable mechanical properties.[180, 181] The bioinks used in 3D bioprinting are categorized into two main groups: cell-scaffold-based approach and scaffold-free cell-based approach.[180, 182] The cell-scaffold-based method uses a bioink that contains biodegradable biomaterial and living cells, and after the bioprinting process, biodegradation begins, and the cells grow to occupy the vacant space. In contrast, the scaffold-free cell-based technique bioprints the living cells directly, in a manner similar to normal embryonic growth. The source of cells used in bioinks also varies. The most commonly used cell sources for OOC applications include stem cells, primary cells, and immortal cells.
2.3.1 Stem cells
Stem cells can sustain themselves through self-renewal, with the ability to become mature cells of various tissues through differentiation, and the capability to repair damaged parts, leading to the development of many different cell types, from heart cells to muscle cells to brain cells.[183-185] The formation of at least one identical daughter cell is the fundamental property that distinguishes stem cells from other body cells; for example, muscle cells, blood cells, and nerve cells cannot divide and self-replicate.[186] The main types of adult stem cells are: (i) hematopoietic stem cells (blood stem cells), (ii) MSCs, (iii) neural stem cells, (iv) epithelial stem cells, and (v) skin stem cells.[187] Although stem cells have advanced regenerative medicine and transplantation techniques, their proper isolation from the body is challenging.[187]
2.3.2 Primary cells
Primary cells are specifically defined as cells that are isolated directly from living organs or tissues and then used immediately.[188] Unlike stem cell and progenitor populations, primary cells cannot divide indefinitely, and the main reason to use primary cells lies in their highly specific nature and efficient mimicry of the biological properties of mature tissues, resulting from their high degree of patient-dependent sample heterogeneity.[189, 190] Primary cells can be used for various purposes, such as on-chip platforms of kidney, liver, neurovascular units, lung, and heart,[191-195] as well as drug resistance testing[196] and drug delivery studies,[197] because primary cells are isolated directly from the tissues or organs and better mimic the in vivo system (although these platforms were not necessarily 3D bioprinted, they can demonstrate the potency of primary cells for future applications in 3D bioprinting of OOCs).[197] Moreover, tissue biopsies have the advantage of preserving 3D natural organ-specific ECM, which is lacking in 2D cultures and many 3D cultures.[198] On the other hand, cell quality is inconsistent in primary cells samples since cell properties may differ between different collection and culture conditions.[5, 198] Furthermore, due to the lack of an effective perfusion system for waste disposal and nutrition supply, a reduction in the functionality of primary cells is conceivable after being removed from the natural environment of the organ. This phenomenon is well observed in primary human brain endothelial cells (ECs), which cannot form a tight barrier in vitro,[185, 199] and in primary hepatocytes, which show decreased enzymatic activity and albumin secretion in vitro if not properly conditioned.[77] Overall, the main challenges with primary cells are limited supply, donor-specific variability, and inability to proliferate indefinitely.
2.3.3 Immortal cells
Immortal cell lines are defined as cell culture systems with the ability to replicate indefinitely in a repeated process.[200, 201] Cell lines arise from cultures of primary cells. After direct initiation from the cells, tissues, or organs of animals or humans, albeit outside their natural environment but under controlled conditions, the lines are grown within a few days to be used in experiments. Cell lines have been used in OOC platforms, including placenta, liver,[202, 203] and multiorgan chip systems, such as combinations of skin, bone marrow, liver, kidney, adipose tissue, gastrointestinal tract, and lung (although these platforms were not necessarily 3D bioprinted, they can demonstrate the potency of immortal cells for future applications in 3D bioprinting of OOCs).[204, 205] In addition, human cancer-derived cell lines are the basic laboratory sources for cancer studies, ranging from its biology to testing therapeutic systems.[206-209] In addition, ECs are promising cells for OOC applications. Since OOC systems enable perfusion of cell culture and emulation of vasculature models, ECs can play an important role by modulating vascular permeability.[210] For instance, in the simulation of acute and chronic inflammation (i.e., hyperglycemia), ECs can regulate the permeability to allow a higher flux of immune cells, which facilitates the study of effects of immune cell intensity in the case of inflammation,[210, 211] highlighting the potency of ECs to be used as a cell source in OOC platforms. However, due to the induction of overexpression of proteins involved in specific toxicity-related pathways, immortal cells have limited applicability for toxicity testing.[212] Additionally, although immortal cells provide more reproducible results compared with primary cells because they have a more homogeneous population, immortal cell lines possess less patient specificity compared with that of tissue biopsies, stem cells, and primary cells, which limits the applicability of immortal cells for disease modeling.[198, 212] Thus, despite being widely used for OOC studies, immortal cell lines face the challenge of adequately emulating the natural physiology of human tissues.
3 APPLICATIONS
This section reviews the most recent biomedical applications of 3D-bioprinted OOC platforms.
3.1 Heart and vessels
The engineering of cardiac tissues and organ models remains a great challenge, due to the special structure of the native myocardium, together with the need to integrate blood vessels, which adds to the complexity. Although in vivo models provide the appropriate environment in terms of physiology and biology, an in vitro surrogate is being introduced for research on tissue development and its functionalities through the advancement of 3D bioprinting technology, which is a reproducible and scalable fabrication methodology with precise 3D control compared with conventional tissue fabrication methods.[75, 213] Moreover, scaffold-free 3D bioprinting can be integrated with conventional 3D tissue engineering methods to obtain a more realistic functional heart and advance science in the treatment of cardiovascular diseases.[213, 214]
In this context, 3D bioprinting has been used to produce endothelialized myocardium.[76] Using a multicomponent bioink and microfluidic technology, ECs were bioprinted directly into microfibrous hydrogel scaffolds. In combination with a specially designed microfluidic perfusion bioreactor, the resulting endothelialized myocardium-on-a-chip system was espoused to demonstrate the cardiovascular toxicity of pharmaceutical compounds. Such a strategy could be applied to human cardiomyocytes derived from induced pluripotent stem cells to construct endothelialized human myocardium. The final diameter of the resulting microfibers after extrusion was 150 μm. Moreover, human umbilical vein ECs (HUVECs), homogeneously distributed after bioprinting, gradually formed a confluent endothelial layer around the microfibers (in 14 days), which resembled the pattern of blood vessel walls. It was also reported that the density of the adherent cardiomyocytes, immediately after bioprinting, was independent of the aspect ratios of the unit grid of the scaffold. It was indicated that perfusion of the bioreactor at a rate of 50 μL/min reduced the number of dead bioprinted cardiac microtissues by approximately 400%. Doxorubicin, an anticancer drug, was used for drug screening assays, resulting in a 70.5 and 1.62% (near to 0 bpm) decrease in cardiomyocytes beating rate, 6 days after exposure to 10 and 100 μM of doxorubicin, respectively, whereas control endothelialized myocardial organoids maintained 88.3% beating rate, highlighting the efficacy of the proposed heart-on-chip platform for drug analysis.[76]
To mimic blood flow and model the in vivo structure of the vessel, GelMA was used to bioprint a 3D vessel-on-a-chip platform with ECs and smooth muscle cells (SMCs) on a microfluidic chip with cell viability of ∼90% (94, 91, and 88% at 1, 4, and 7 days post-printing, respectively).[215] The microfluidic chip was carved out of polymethylmethacrylate (PMMA) using a high-precision computer numerical control engraving machine. Compared with the conventional culture platforms, the EC-SMC coculture chip model resulted in a greater upregulation of alpha smooth muscle actin (αSMA) and SM22 protein expressions of the SMCs, and maintenance of the SMC contractile phenotype, mimicking the microenvironment of the natural vasculature under fluid flow conditions. This method enabled the establishment of an in vitro vascular model for physiologically relevant studies and the exploration of pathological processes in the vessel wall.[215]
Thrombosis and its complications are one of the major causes of morbidity and mortality from cardiovascular disease, bringing about more deaths than trauma and cancer combined.[216] To study thrombosis more effectively, an in vitro thrombosis-on-a-chip was fabricated using sacrificial 3D bioprinting technology to explore potential therapies and understand cellular interactions.[216] The sacrificial layer was printed as a scaffold for UV curation of GelMA hydrogel to form hollow microchannels of the model, which were populated with HUVECs, into which human whole blood was infused immediately after thrombosis activation. Crosslinking of the hydrogel with longer UV exposure times resulted in matrice with higher moduli (0.8 kPa for 25 s, and 0.65 kPa for 10 s). However, increasing the exposure time to 20 and 25 s amplified the death rate of the embedded cells in the GelMA hydrogels and limited the spread of the encapsulated fibroblasts, whereas 10 and 15 s of UV exposure caused little damage to the cells (>80% cell viability). The platform was able to mimic thrombus within 10 min after application of 0.1 M of CaCl2 in Dulbecco's phosphate-buffered saline. The thrombolysis test was performed by forming an artificial thrombosis on this setup to investigate the clot-dissolution ability of the tissue plasminogen activator (tPA), which showed the efficacy of tPA treatment within 2 h. The platform allowed the simulation of blood flow in the human leg (the common site of thrombosis formation) for burst pressure (∼0.16 kPa) with different flow rates (ranging from 0.6 to 3 mL/h) and velocities (ranging from 0.19 and 0.54 mm/s). Potentially, specific patient-derived cells can be used in the fabrication process to study fibrosis pathology and personalized medicine for vascular fibrotic diseases.[216]
A multimaterial cardiac microphysiological device was developed on a chip for direct noninvasive electronic readout of contractile stresses to perform dose–response studies of drugs that influence contraction strength or beat rate.[217] The wells were incubated with fibronectin (FN) solution in PBS for 1 h, following well seeding by either human-induced pluripotent stem cell-derived cardiomyocytes (hiPS-CMs) or primary neonatal rat ventricular myocytes. Following culture, the sarcomere packing density (a measure of periodic sarcomere organization) increased from 0.07 to 0.2, and the sarcomere orientational order parameter increased from 0.11 to 0.32, from day 2 to day 14, demonstrating that hiPS-CMs undergo myofibrillogenesis and sarcomerogenesis during culture. Besides, 4 weeks after culture, a significant increase in sarcomere length from 1.7 to 1.8 μm was observed (from day 14 to day 28), indicating a more mature tissue. This platform showed the potency of on-chip platforms for the engineering of laminar cardiac tissues with a range of ordered architectures.[217]
3.2 Brain and BBB
Brain tumors exhibit a dynamic complexity consisting of different cell types.[218] Glioma is the most aggressive brain tumor known to be responsible for approximately 48.3% of malignant brain tumors and other central nervous system (CNS) tumors[219] and causes nearly 3% of cancer-related deaths annually.[220, 221] Although mouse models are widely used for large-scale genomic analyzes and the study of biological mechanisms of tumorigenesis, genetic differences between humans and mice lead to an erroneous recapitulation of human pathophysiology. Despite advances in multimodal medical treatment, ∼70% of grade-II glioma tumors still progress to grades III and IV, which are lethal within 12–14 months.[222] To promote patient survival, low-cost and accurate platforms are needed to critically analyze the molecular biology of glioma cells, as well as their interactions with the immune system and potential drug candidates.[223] 3D bioprinting is one of the emerging methods that can be used to fabricate brain-on-chips,[224] especially for glioma modeling,[104] along with other applications such as developing patient-specific therapeutics[225] and studying drug resistance of brain tumor cells,[226] which is also important for developing methods to prevent tumors recurrence.[227, 228] In addition, some specific methods such as lab-on-a-printer technology can also be used for 3D bioprinting of neural tissues.[229]
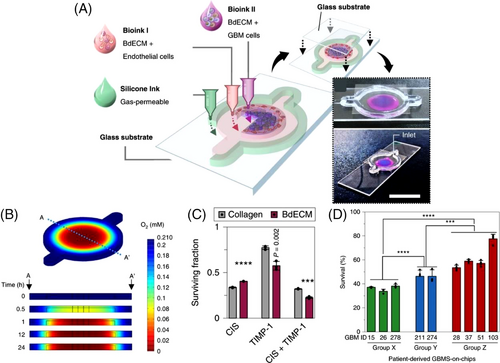
Glioblastoma (GBM), a grade-IV astrocytoma, is a rapidly growing and aggressive brain tumor. Identification of patient-specific drug sensitivity and the development of more beneficial personalized cancer therapies can be achieved by ex vivo GBM-on-a-chip models.[230, 231] To bioprint an ex vivo platform onto a glass substrate, silicon ink (to print the chamber wall) and GBM cells as well as vascular cells (HUVECs) were embedded into brain-decellularized ECM (BdECM) or collagen gel to determine patient-specific therapy resistances and to investigate drug combinations for a more effective tumor-killing performance of treatments (Figure 3A).[232] This platform was equipped with an oxygen gradient-generating system to create an in vivo-like biomimetic oxygen gradient in the chip (Figure 3B). The printing process was performed using an in-house 3D printer (with a tapered nozzle) to print silicone ink where the custom bioink was pushed down through a flat needle at a speed of 500 nl/s. Human GBM cells were embedded in the BdECM and collagen gels and showed >90% cell viability in both, with a higher cell proliferation rate in BdECM after 10 days (Figure 3C). Moreover, higher expression levels of genes encoding proangiogenic factors (interleukin 8 (IL-8) and vascular endothelial growth factor A) and ECM-remodeling proteins (matrix metallopeptidase 9 (MMP9), MMP2, MMP1, FN, and protein tyrosine kinase 2) were detected for the BdECM gel compared with the collagen gel after 3 days. After 14 days, the cluster of differentiation 31-positive (CD31+) ECs produced more active tubule networks in the BdECM gel compared with than in the collagen gel, while the BdECM gel also showed superior capacity in terms of angiogenesis of HUVECs. Adjuvant concurrent chemoradiation therapy (CCRT) with temozolomide (TMZ), the most common therapy for GBM, was applied on the chips prepared from the patients’ GBM cells. According to the current clinical approaches, patients in group X had the highest survival rate, and those in group Z had the lowest survival rate. Following drug treatment, the chips made from group X's cells showed <40% cancer cell viability in response to drug treatment, whereas the group Z GBM-on-chip models showed >60% cancer cell viability, indicating the ability of this platform to accurately predict malignancy grade (Figure 3D).[232]
A custom-made inkjet bioprinter was used to bioprint sodium alginate as a matrix for encapsulating cells (HepG2 and human glioma cell line (U251)) onto PDMS microchannels (prepared by soft lithography) for drug stimulation and diffusion experiments.[130] It was highlighted that higher concentration and viscosity of the hydrogel resulted in a stronger hydrogel structure, while making the bioprinting process more difficult (droplets did not detach from the nozzle). Using larger nozzles facilitated the printing of viscous materials while sacrificing ultimate print resolution. In addition, applying a higher voltage solved the detachment issue by increasing the force generated by the piezoelectric material at the nozzle head, while compromising cell viability. Therefore, there was a trade-off between the viscosity of the bioprinting material, the applied voltage, and the nozzle size. In the study, a 0.5% concentration of alginate and a voltage of 40 V were chosen to produce the proposed chip using an inkjet bioprinter. Higher hydrophilicity of the glass substrate improved cell adhesion to the substrate and print resolution. By optimizing the hydrophilicity, a resolution of 400–1000 μm was achieved. To evaluate the drug modeling capability, the prodrug Tegafur was used as a model drug that can be metabolized by liver cells into the anticancer drug 5-fluorouracil, which acts on the U251 cells (glioma cells). Tegafur concentrations of 100 and 1000 μM were applied and successfully reduced the cancer cell viability of U251 cells to ∼80 and ∼50%, respectively. In addition, the OOC platform clarified that Tegafur inhibits cancer cell proliferation and is effective on U251 cells in the copresence of HepG2 cells, whereas it is not effective in the absence of HepG2.[130]
The BBB—the barrier of ECs with high selectivity that shields the brain and CNS from solutes in circulating blood—is one of the challenges in delivering therapeutics to brain cancer cells, especially the GBM tumor foci,[233, 234] and hence modeling the BBB will be useful for cancer studies. The TPP technology was used to bioprint a 1:1 scale microfluidic BBB chip composed of porous microcapillaries.[235] Inspired by brain capillaries, the microtubes of the system were designed and bEnd.3 ECs from mouse brain and U87 GBM cells were used for seeding inside the chip. Numerical analyzes including intensity plots of fluid velocity and axial velocity profiles were used to investigate the proposed model in terms of the flow occurring, which demonstrated a uniform flow rate inside the channels similar to real physiological conditions. Confocal laser scanning microscopy 3D imaging of the model depicted that the bEnd.3 ECs were able to efficiently cover the tubular structures after 3 days of culturing. Furthermore, high-magnification scanning electron microscopy imaging of the setup provided qualitative evidence that the ECs almost completely covered the pores. By modifying parameters such as the diameters of the pores and microcapillaries, the pore density, and the length of the porous segment, the platform could be used as an in vitro model for a variety of studies such as drug screening.
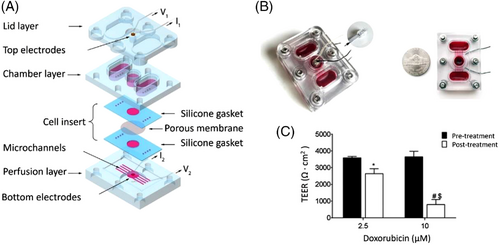
Screening of drug permeability in the BBB was also investigated using a microfluidic BBB-on-a-chip fabricated with an SLA 3D printer.[81] The fabricated pumpless chip consisted of three layers (lid, chamber, and perfusion), as shown in Figure 4A, and was able to mimic the in vivo properties of the BBB, making it a suitable platform for in vitro investigation of drug permeability. Brain microvascular ECs, derived from hiPSCs, were used for this study. These cells were cocultured for up to 10 days with rat primary astrocytes in a microfluidic system (Figure 4B), designed based on the residence time of blood in human brain tissues to ensure physiological transfer of nutrients and exogenous substances in a realistic manner around the BMECs without the need for pumps or tubing in the microfluidic system. The amounts of trans-endothelial electrical resistance (TEER) in the studied system were similar to the in vivo values. The TEER value was ∼3500 Ω · cm2 and decreased to as low as ∼800 Ω · cm2 after 24 h treatment with 0–10 μm doxorubicin (Figure 4C), demonstrating the achievement of significant barrier integrity in the proposed microfluidic system.
A 3D vascularized neural construct was developed for in vitro reconstitution of BBB function.[236] 3D interconnected blood vessels were simulated by seeding ECs within the channels of the network, along with other types of cells, including neurons, astrocytes, and pericytes, in a collagen matrix, wrapping the vasculature network to derive a vascularized neural construct that recapitulates in vivo BBB function. A dopamine/collagen coating was used on the surface of the PCL/PLGA tube for the functionalization, increasing the surface hydrophilicity (water contact angle reduced from 104.7° to 33.8°) for improved cell adhesion and growth. Successful maturation of the culture was examined with the TEER test (raising from ∼40 to ∼120 Ω · cm2 after 8 days), indicating a matured vascularization process for restricting compound transportation. The barrier capacity of the developed culture was tested by measuring the leakage of fluorescently labeled dextran of different molecular weights (10 kDa (2 nm), 70 kDa (11 nm), and 150 kDa (15 nm)). The cross-endothelial spreading in the peri-tubule region was 13% for 10 kDa (2 nm) dextran after 15 min, which was 40% for even larger 70 kDa (11 nm) dextran in the nonendothelialized case, showing the favorable impermeability of the developed model. Cell viability of ∼80% was recorded for the perfused model, compared with 50% in the nonperfused chip. Moreover, compared with nonvascularized tissue, neuronal growth in the vascularized tissue construct was longer with continuous neurite extension (perfused: 175 μm; nonperfused: 117 μm), underlining the vital supporting role of the vasculature system in the reconstruction of functional in vitro models.[236]
In another study, a fluorescent molecule (fluorescein isothiocyanate (FITC)-dextran)-based test was used to investigate receptor-mediated transcytosis for screening purposes (receptor-mediated transcytosis is one of the major routes for drug delivery of large molecules into the brain).[237] In this regard, immortalized human brain ECs, astrocytes, and pericytes were used to form a blood microvessel, which was grown adjacent to ECM gel, and a third channel to insert astrocytes and pericytes. The lumen of the endothelial vessel of the model was perfused with a controlled antibody or an antitransferrin receptor antibody in order to test antibody transcytosis. The barrier function/integrity was assessed using fluorescent barrier assay in which the leakage of a 0.1-mg/mL 20 kDa FITC-dextran dye (with a hydrodynamic radius of 3 nm) from the microvessel into the adjacent gel channel was measured by the acquisition of fluorescent images over time (using an ImageXpress XLS Micro HCI system (molecular devices)). Penetration of the antibody targeting the human transferrin receptor (MEM-189) was markedly higher (permeability of 2.9 × 10–5 cm/min) than penetration of the control antibody (1.6 × 10–5 cm/min), showing the potency of the developed OOC platform to retain all fluorescent dye within the vessel.[237]
3.3 Lung and airway
According to the World Health Organization, chronic respiratory diseases (CRDs) (i.e., lung and airways diseases) are currently not completely curable. Nonetheless, drug treatments can control symptoms and relieve breathlessness by dilating the airways to improve patients’ quality of life.[238] CRDs include asthma (235 million people suffer from it, mostly affecting children, 14% of children worldwide[238]), lung cancer (the leading cause of cancer-related deaths (1.8 million deaths) with 2.2 million newly diagnosed cases in 2020[239]), chronic obstructive pulmonary disease (causing 3.23 million deaths in 2019[240]), pulmonary hypertension (occurs in ∼1% of the world's population), and occupational lung disease.[241] Severe air pollution, smoking, occupational chemicals, and childhood lower respiratory tract infections are important risk factors contributing to CRDs. Lung-on-a-chip platforms allow correlation of various risk factors (e.g., genetics and chemicals) with the likelihood of developing CRDs, as well as real-time analysis of drug responses to patient-derived cells to achieve more effective personalized medicine and treatment. In recent decades, there have been attempts to fabricate airway-on-a-chip platforms.[242, 243] For instance, a porous PDMS membrane was used to simulate the alveolar–capillary interface of a human lung on a chip to model the disease of pulmonary edema and to study organ-level responses to bacterial and inflammatory cytokines.[243] However, this model faced the challenge of stably reproducing the dynamic structures of native 3D vascular networks over the long term. In addition, most conventional OOC platforms required manual fabrication steps that limited reproducibility.[243]
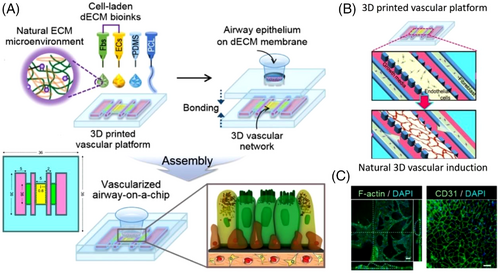
An airway-on-a-chip, with an integrated natural vascular network, was 3D bioprinted in vitro using decellularized ECM bioink derived from porcine tracheal mucosa-derived dECM (tmdECM), as bioink, on a PCL frame to simulate respiratory disorders, such as allergen-induced asthma exacerbation and asthmatic airway inflammation (Figures 5A and 5B).[242] The tmdECM hydrogel was prepared from the porcine trachea, where its rheological properties were studied before and after incubation and compared with collagen I (Col-1) and Matrigel using an advanced volumetric expansion system. The vascular platform, including PCL, EC bioink, and lung fibroblast bioink, was bioprinted using an in-house pneumatic extrusion-based 3D bioprinter. The fabricated platform with a mixture of tmdECM and Matrigel was able to form an extensive vascular network compared with Matrigel alone (Figure 5C). Self-assembly of ECs in tmdECM resulted in an interconnected vascular network after 7 days that was stable for up to 3 months under static culture conditions in vitro. The VP was also useful for monitoring tissue homeostasis, as its TEER (an indicator of cellular barrier integrity in the airway epithelium) was within the range of a healthy human tracheal epithelium. The vascularized airway-on-a-chip (VA-OC) was treated with IL-13, a cytokine that induces allergic asthma in human airway epithelium, to test the chip as an in vitro asthma model. Without affecting cell viability, treatment triggered the production of hRANTES (human regulated upon activation, normal T cell expressed and presumably secreted) and human tumor necrosis factor-α, the major inflammatory cytokines in asthma, at rates of 13.327 and 15.479 pg/mL, respectively, demonstrating the reproducibility of pathological interactions between the airway epithelium and vascular network using the VA-OC platform.[242]
In order to recapitulate key features of the lower respiratory airways on a chip (specifically the blood vessel–interstitium fibroblast–epithelial microenvironment), a thin film (polyester track-etched (PETE) or vitrified collagen) membrane and a microvascular platform were integrated to grow epithelium at an air–liquid interface.[244] The macrophysiologic device was comprised of a commercially available bottomless 96-well plate, PETE or vitrified collagen membranes, a 3D printed airway layer, and a photolithography-based vascular layer. Normal human lung fibroblasts (NHLF), in a fibrin gel, were seeded into the two outer channels, which provided the needed cytokine gradient to direct vasculogenesis of the HUVEC fibrin gel culture in the central channel. The PDMS layer was designed for epithelial cell growth and providing the open ports to seed and feed both culture layers. For NHLF-B devices, the αSMA median area expanded from 0.4% (for the untreated group) to 1.9 and 1.0% for pirfenidone and TGF-β1 treatment, respectively. In addition, the fluorescent area of the migrated neutrophils across the vasculature was higher in the PBS cystic fibrosis human bronchial/epithelial cells (PBS CF-HBE) devices compared with the normal human small airway epithelial cells, confirming that the device accurately modeled how CF epithelial cells increase neutrophil migration compared with normal epithelial cells. Despite the successful implementation of the model, it was suffering from a lack of expansion/contraction of the membrane to replicate the cyclic mechanical strain of breathing, and a lack of dynamic movement of media in the endothelial compartment.[244]
3.4 Liver
The liver is the main organ for protein synthesis, bile acid production, biotransformation of drugs, detoxification, and filtration in the body.[245] Since administered drugs enter the liver via the bloodstream, drug-induced liver injury is prevalent, highlighting the unmet need for the development of in vitro liver models for preclinical and clinical drug screening. Liver-on-a-chip methods are beneficial for understanding the liver function and disease impairment, studying the influences of dietary and cosmetics supplements on the human organ, screening foodborne pathogens/diseases, and monitoring drug toxicity/efficacy.[29, 246] Despite the high proliferation and regeneration capacity in conventional 2D and 3D cultures, the functionality (e.g., enzymatic activities and albumin secretion) of in vitro hepatocytes from human liver biopsies decreases after being removed from the natural environment of the liver (within 1 week)[247] because of the lack of an efficient perfusion system for waste product removal and nutrient delivery. On the other hand, liver cells can be cultured on a chip for a longer period of time in that liver-on-a-chip platforms provide continuous perfusion through microfluidic channels.[77] Cell sources for liver-on-a-chip platforms include stem cell-derived hepatocytes (a consistent source of hepatocytes, but requiring specific induction factors and facing difficulties to manipulate), liver-derived cell lines (easy to manipulate and endowed with unlimited lifespan, but with rapid loss of expression of liver-specific transporters/enzymes and inaccurate drug response), and primary human hepatocytes (possess intrinsic properties of the liver, but are unsuitable for long-term culture, costly, and difficult to isolate).[246, 248]
For analysis of human HepG2/C3A spheroids for drug toxicity over the long term (30 days), a continuously perfused, syringe pump-operated liver-on-a-chip setup was bioprinted (Figures 6A and 6B).[77] PDMS was cast around a laser-cut PMMA mold to form a set of channels and three chambers. Unlike most PDMS-based microfluidic chips, where the different layers are permanently sealed by plasma, this setup was sealed with screws and nuts to prevent compound leakage, facilitating disassembly of the chip at any stage of the experiment for direct cell culture analysis. Fifteen seconds of UV light exposure (850 mW) was used to crosslink the bioink that were consisted of GelMA and HepG2/C3A spheroids (191 ± 10 μm). Drug simulation was performed with 15-mM acetaminophen (APAP) to induce a toxic response in the hepatic culture, giving results comparable to those obtained in animal experiments. The cell number was increased tenfold after 30 days of cell culture, which is a common cell density range for hydrogel encapsulation. By measuring the secretion of biomarkers (albumin, ceruloplasmin, alpha-1 antitrypsin (A1AT), and transferrin), the hepatic function of the proposed setup was examined.[77] Similar setups were proposed, which were able to efficiently recapitulate key hepatic tissue cell types, such as hepatocytes, Kupffer, endothelial, and stellate cells, the distribution of various ECM-like biomaterials, and their ratios.[249]
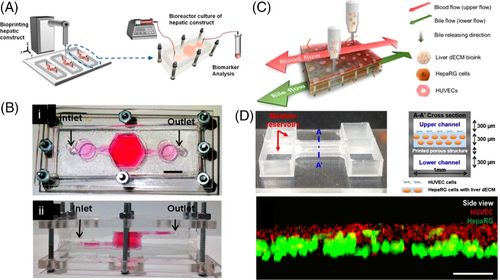
In another study, a gravity-induced (i.e., pumpless) liver-on-a-chip setup (0.03 × 0.1 × 1 mm3) was 3D bioprinted in a single step with perfusion capability (at a flow rate of 25 μL/min). The chip consisted of vascular/biliary channels for waste removal (lower biliary channel) and supplying nutrients (upper vascular channel), two reservoirs to ensure a continuous supply of compounds within the channels, and a 3D liver-decellularized ECM (dECM) environment for the cells (Figures 6C and 6D).[245] Structural printing was performed under 660 kPa and 110°C conditions using poly(ethylene/vinyl acetate) and sterilized transparent PMMA as structural material and printing substrate, respectively. Cells were bioprinted using gelatin and liver dECM bioinks. The precision of cell bioprinting was evaluated by acquiring images of the cell layer, showing that HUVECs were accurately placed over the human hepatoma (HepaRG) cell-laden liver dECM bioink (Figure 6D). To examine the ability of this platform in emulating the basic function of liver cells, albumin/urea secretion in 2D culture was compared with that of the liver-on-a-chip platform. This demonstrated a higher level of secreted urea and albumin in the 3D bioprinted model, whereas the secretion level in the 2D model continuously decreased. Furthermore, the drug response of the model was analyzed by applying 5-mM APAP and measuring albumin secretion, reporting a higher drug sensitivity and reactivity in the 3D-bioprinted platform compared with the 2D model. The simulation of the liver-like microenvironment and the presence of multiple cell types and biliary channels were the main reasons for promoting the functionality of this setup.[245]
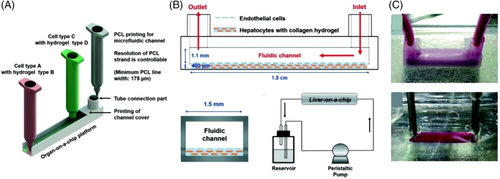
Despite the considerable attention given to OOC platforms, this technology still suffers from protein absorption, challenges in forming different types of ECM environments for cell–ECM interactions, and poor selectivity of different cell types in the presence of spatial heterogeneity, mainly due to conventional fabrication methods (e.g., soft lithography, microcontact printing, and molding). To overcome these limitations, PCL may be a practical candidate. PCL is a nontoxic, biodegradable, and biocompatible polyester with a low melting point (∼60°C), resulting in high cell viability for 3D bioprinting. A single-step 3D bioprinting approach was introduced, without a secondary cell seeding process, to produce a perfused liver-on-a-chip platform with PCL-based 3D printed microfluidic channels (Figures 7A and 7B).[8] PCL, as microfluidic chip material, and hydrogels containing encapsulated HUVEC and HepG2 cell lines were printed using a pneumatically activated nozzle 3D printer with a channel size of 1.5 mm × 1.5 mm × 15 mm and minimum line widths of 175 μm. The protein absorption rates of the PCL-based bioprinted 3D channels (protein absorption depth in the channel wall: 50 μm, absorbed dye in the channel: 3.4% of inlet concentration) were compared with those of soft lithographic PDMS-based channels (absorption depth in the channel wall: 400 μm; absorbed dye in channel: 10.5% of inlet concentration), confirming the better performance of PCL-based channels on heterotypic cell types and biomaterials for more accurate drug screening. However, PCL-based microfluidic channels exhibited lower optical transparency compared with PDMS channels (Figure 7C), which is one of the important features for cell proliferation/viability rate assessment and real-time analysis of cell interactions and drug responses.[8]
3.5 Gut
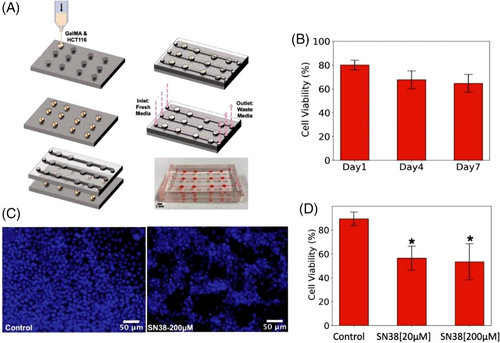
Colorectal cancer, as the second leading cause of cancer-related death in the United States, was diagnosed in 147,950 individuals, in 2020, resulting in 53,200 deaths.[250] To find more effective therapies, an OOC-like platform was implemented on a PDMS-based microfluidic chip (molds made of Pluronic F127) through a microextrusion 3D bioprinter to study colorectal carcinoma (Figure 8A).[2] The structure consisted of channels (allowing physiological fluid flow onto the cell constructs) with width, length, and depth of 800 μm, 30 mm, and 300 μm, respectively, and concave wells (to hold the 3D-HCT116 cell constructs) with a diameter of 1.5 mm. Bioink, consisting of alginate and nanofibrillar cellulose, was mixed with HCT116 cells (in a 10:1 ratio) and bioprinted onto concave wells through a 410-μm nozzle at 4 kPa. The presence of bioink not only provided an analogous microenvironment to native ECM in the human body, but also protected the cells during bioprinting through the nozzle, with enhanced cell viability. The viability of the constructs was examined using cell nuclear staining and a fluorescence microscope. The cell viability of the 3D spherical cultures was measured to be 80.1, 67.8, and 64.7% at 1, 4, and 7 days after bioprinting, respectively (Figure 8B). Drug toxicity assays were performed with 7-ethyl-10-hydroxycamptothecin (SN-38), used in colon cancer, on the chip with three HTC116 construct arrays with separate microchannels. While the control line had a cell viability of 90%, application of 20 and 200-μM SN38 resulted in cell viability of 57 and 48%, respectively, 48 h after drug treatment (Figures 8C and 8D).[2] Additionally, the potential impacts of bioprinting parameters were also scrutinized. Although smaller nozzle sizes resulted in better 3D bioprinting resolutions, reducing the nozzle dimensions from 410 to 200 μm increased the pressure required for bioprinting from 90 to 110 kPa, which augments the shear stress that cells are subjected to during the bioprinting process, which may have a negative effect on the cell viability rate. Besides, changing the nozzle shape from a conical to a needle-shaped nozzle increased the minimum bioprinting pressure by ∼120% for the same size. Moreover, the prevention of entrapped air bubbles is a challenging problem in 3D bioprinters, which can be addressed by optimizing the bioprinting characteristic, such as bioprinting pressure as well as speed, nozzle size, composition, and proportion of constituents in the bioink.[2]
3.6 Renal system
The human kidney, filtering nearly 180 L of blood daily, is susceptible to blood-borne diseases and drug-induced injuries.[251] Chronic kidney disease (CKD) refers to the malfunction of the kidney in filtering blood. As the ninth leading cause of death in the United States, the economic burden of kidney diseases was roughly $118.4 billion in the United States, in 2018, including diagnosis, drug treatment, kidney transplantation, and dialysis.[252] Hypertension, diabetes, a family history of kidney failure, and heart disease are the main risk factors for developing kidney disease—three out of four cases of kidney failure are due to diabetes or hypertension.[253] CKD can result in kidney failure, heart disease, anemia (i.e., low red blood cell count), infection, low calcium, high potassium, and high phosphorus levels in the blood. It is estimated that 5.24 million people will require dialysis by 2030, highlighting the urgent need to develop effective treatments for kidney disease.[254] Early kidney models, cultured on hollow fibers or biomimetic basement membrane coatings, were able to maintain a differentiated state and self-organization of cells.[255-260] Later, more complex 3D microenvironments were proposed, such as differentiated proximal tubule cells in thin gels[261, 262] and induced pluripotent stem cell-derived kidney organoids with nephronal properties.[263-266] Kidney-on-a-chip platforms mimic the structure of the human kidney and its functions, such as reabsorption, and thus presenting a promising tool for disease simulation, drug screening, and personalized therapies. Current kidney-on-a-chip platforms face the challenge of emulating complex 3D structures, drug responses, and tissue functions, such as reabsorption[251]—the reentry of amino acids, potassium, glucose, and water filtered during glomerular filtration into the bloodstream via passive concentration gradient-based transfer in the proximal convoluted tubule of the nephron.[267] Despite achieving complex 3D structures, the kidney organoids faced challenges in longevity, perfusion under physiological shear stress, perfusate collection, and analysis similar to 3D convoluted and open luminal architecture.[72, 268] The 3D bioprinting technology can produce complex luminal tissue architectures, overcoming the current limitations of kidney models.
Human renal proximal tubules were printed in 3D on a chip with a perfusable open lumen architecture, confined by proximal tubule epithelial cells (PTECs), and analogous physiological shear stresses that can be maintained viable for 2 months.[72] Both the silicon gasket and the fugitive ink (38 wt% Pluronic F127 and 100 U/mL of thrombin in deionized ultra-filtered water), for the tubular hollows, were printed onto a glass slide using a pneumatically activated nozzle extrusion 3D printer. The fugitive ink was then evacuated, leaving hollows inside the ECM for proximal tubules. PTECs, designed for proliferation, were then seeded to form the tubules and cultured using a continuous supply of cell media at shear stresses between 0.1 and 0.5 dynes/cm2. Using transmission electron microscopy, it was observed that the growth height of the 3D perfused tissue cells was 14.1 μm, which was 100% more than the growth height of the nonperfused planner platforms, 40% more than that of the perfused 2D culture, and closer to healthy human proximal tubules (20.3 μm). Furthermore, the average microvilli length on 3D perfused tissue (1.24 μm) was also 200% longer than that of 2D nonperfused culture, ∼40% more than that of 2D perfused platforms, and was closer to in vivo values (2.89 μm), demonstrating the superiority of 3D-bioprinted platforms, over other conventional cultures, in simulating in vivo-like conditions. Albumin uptake, a crucial indicator of homeostasis, was observed to be higher in 3D tissue than in 2D. Moreover, the expression of megalin, one of the transporters for albumin, was the highest in 3D-printed culture among 2D cultures. Finally, for drug tests, the effects of cyclosporine A (CysA), a harmful nephrotoxin for proximal tubule cells, were monitored by perfusing it into the cell culture at varying concentrations. The epithelial barrier permeability was increased sixfold and fourfold by exposure to 100 and 500 μM of CysA, respectively.[72]
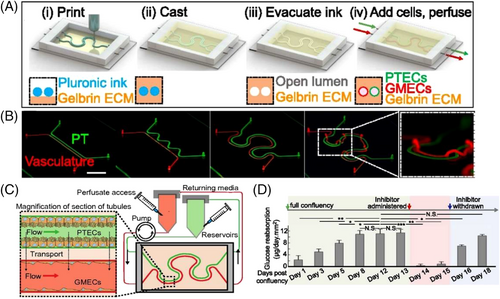
In another study, using a modified ECM, an on-chip perfusable 3D human vascularized proximal tubules tissue was 3D-printed using a tubular–vascular exchange to simulate renal reabsorption (Figures 9A–9C).[251] Although a minimum channel diameter of 20 μm was printable, seeding the high density of cells into channels smaller than 200 μm was challenging. It was reported that reducing the gelatin-to-fibrin ratio from 7.5 to 0.4, in the modified ECM, reduced the time required to obtain a confluent epithelium (fourfold reduction from ∼21 days to ∼4 days). Besides, a drug screening assay was performed on the 3D-printed kidney-on-chip using dapagliflozin, a glucose reabsorption inhibitor. Application of dapagliflozin resulted in ∼98% reduction in glucose reabsorption, highlighting the specificity and regulability of the developed on-chip model for longitudinal studies of drug responses (Figure 9C). Hyperglycemia, a sign of diabetes and a risk factor for vascular disease, was also modeled on this platform by circulating a perfusate with a fourfold higher glucose level and monitoring EC damage.[251]
3.7 Breast
Cases of breast cancer, the most commonly diagnosed cancer in women, increased by 0.3% per year from 2012 to 2016.[269] However, the mortality rate of breast cancer was reduced by 40% (averting 375,900 deaths) from 1989 to 2017, largely due to the development of effective therapies.[269] Still, a controlled culture of breast cancer cells and administration of drug candidates to monitor the effects of treatments on the cancer cell and surrounding tissue may contribute to more effective therapies. Nonetheless, drug screening and recapitulation of cancer cell growth in an in vivo-like 3D structure are still a challenge in the development of chemotherapy, along with surgery and radiotherapy, as the most common cancer therapy.[17] In this regard, a drug screening system, termed 3D tumor array chip (3D-TAC), was bioprinted with GelMA hydrogel droplets (∼0.1 μL) containing MDA-MB-231 breast tumor cells and evaluated with epirubicin as well as paclitaxel, antitumor drugs, to demonstrate the compatibility of this platform with traditional screening approaches.[17] Although the chip basement was a transparent conductive membrane, the culture chambers consisted of a silicon interlayer and stainless steel. MDA-MB-231 cells, encapsulated in GelMA, were bioprinted onto the conductive membrane using a high-voltage electrohydrodynamic 3D bioprinter through stainless steel nozzles at 2.7 kV and a flow rate of 10 μL/min, and crosslinked with 405-nm light. Despite the high electric field force during the process, the cell viability was above 90%, indicating the high performance of the electrohydrodynamic bioprinting technique. To investigate the proliferation behavior of MDA-MB-231 cells, the nicotinamide adenine dinucleotide (NAD+) examination was tested by the enzyme-linked immunosorbent assay (ELISA) method. The half-maximal inhibitory concentration (IC50) values of the 3D models were 47.63 and 28.83 μM for paclitaxel and epirubicin, respectively, higher than those of the 2D at 46.09 and 27.77 μM, confirming the inhibition of tumor cell proliferation by the applied drugs.[17]
Although the integration of blood vessels into OOC platforms for perfusion is widespread, there are a few reports of the simultaneous implementation of blood and lymphatic vessels in in vitro tumor models. The lymphatic system drains fluid (known as lymph) that has seeped into tissues from blood vessels and returns it to the bloodstream—it plays a key role in the immune system by protecting the body from disease-causing invaders, removing cellular waste, absorbing fats from the digestive tract, and maintaining the body's fluid balance.[270-272] In cancer therapies, the lymphatic drainage system provides a preferential recycling pathway for applied antitumor drugs in vivo and plays a critical role in metastasis, highlighting the importance of developing perfusable OOC platforms with blood and lymphatic vessels.[18, 273-277] A tumor-on-a-chip platform was bioprinted on a microfluidic bioreactor with a dynamic microenvironment consisting of blood and lymphatic vessel pair and MCF-7 breast cancer cells in a 3D hydrogel matrix based on GelMA on a PDMS–PMMA frame (Figures 10A–10F).[18] The vessels were bioprinted using a custom-made bioprinter with a trilayered coaxial extrusion channel featuring different needle sizes. The permeability values of the vessels were measured by observation of FITC–bovine serum albumin and determined to be ∼5.07 × 10–7 cm/s for the blood vessel and ∼1.76 × 10–6 cm/s for the lymphatic vessel, which was comparable to the native in vivo values of vessels (1–8 × 10–7 cm/s for blood vessels, 0.1–5 × 10–6 cm/s for lymphatic vessels[18]). It was also reported that the presence of a second channel (lymphatic vessel) not only caused higher cell viability (Figure 10G) by avoiding drug accumulation in the cell culture, but also accelerated the diffusion rate of FITC compared with the simple perfusion case (blood vessel only) (Figures 10H and 10I). Furthermore, embedding cancer cells in the GelMA slowed the rate of drug or biomolecule (FITC) transfer.[18]

3.8 Bone and cartilage
The study of bone and cartilage models not only helps to understand common bone defects and fractures, but also paves the way for a better understanding of various tumors such as lung, breast, prostate, and melanoma carcinomas, for which the skeletal system of the human body is a chronic site for metastasis of these cancer cells (for instance, in metastatic breast cancer, bone metastasis occurs with a frequency of ∼70%[278]).[279] Although the 5-year survival rate of breast cancer patients, without bone metastasis, can be as high as 98% with early diagnosis and therapy, the occurrence of bone metastasis decreases this rate to 26%, highlighting the importance of understanding the mechanism of bone metastasis and the means of prevention.[278] In addition to cancer, mild bone diseases (e.g., osteoporosis and low bone mass) and subsequent complications affect the quality of life and cause socioeconomic impact. Approximately 43.6 million people over the age of 50 in the United States suffer from bone diseases, resulting in hospitalizations for hip, spine, or wrist fractures with an estimated health care burden of $25 billion in 2025.[280, 281] Common challenges in bone analysis with current models include: the complexity of obtaining antemortem samples of bone metastases, a limited number of ex vivo and in vivo models that effectively emulate bone structure, the physical difficulty of manipulating bone as tissue, and the limitation imposed by examination times, making the real-time monitoring of bone diseases and bone metastases difficult.[278] Recent advances in 3D bioprinting, microfluidics, and OOC science have led to a narrowing of the gap between laboratory constructs and physiological tissues, resulting in the introduction of personalized therapies,[282] a better understanding of the underlying causes of bone diseases, and more effective reconstitution of the cancer microenvironment.[283-287]
Using a desktop 3D printer to create plastic molds followed by replica molding with PDMS, a spontaneous 3D bone-on-a-chip was fabricated to study breast tumor metastasis.[278] As illustrated in Figures 11A–11C, the bone-on-a-chip setup was fabricated based on simultaneous growth dialysis and contained a solution reservoir in the upper part and a cell growth chamber in the lower part. The design of the chip was able to allow long-term growth for up to 30 days, resulting in the formation of a thick mineralized osteoblastic tissue in vitro. To characterize the maturation of osteoblastic tissue on the chip, normalized alkaline phosphatase activity, over 720 h of culture, showed that nitrocellulose-bone-on-a-chip (NC-BC) had the highest activity (Figure 11D). In addition, normalized measurement of insoluble ECM demonstrated that NC-BC collected the highest amount of insoluble ECM (Figure 11E). Etching the glass coverslip with a base solution for a longer period of time (72 h instead of 24 h) increased surface nanoroughness and surface hydrophilicity, which ultimately improved osteoblastic tissue attachment for long-term cultures. In addition, reducing the height of the culture chamber from 5 to 2 mm promoted both mineral and collagen deposition. It was shown that several important features of breast cancer colonization in bone were observed when coculturing metastatic human breast cancer cells with the osteoblastic tissue using the fabricated bone-on-a-chip. For instance, the colony-forming potentials of MDA-MB-231GFP and MDA-MB-231-BRMS1GFP cells were compared, and the former was found to be more likely to form micrometastases (88.5%) than the latter (61.4%).[278]
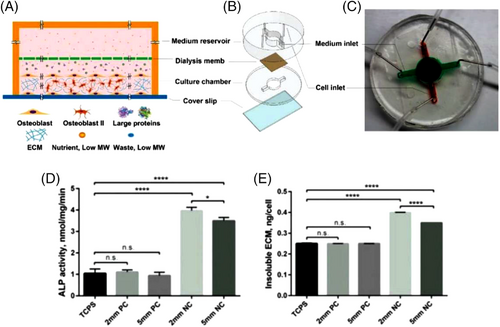
3.9 Skin
In 2018, nearly 1.3 million new cases of skin cancer were diagnosed, with 64,000 reported deaths from nonmelanoma skin cancer. Encouragingly, the mortality rate from melanoma skin cancer has steadily declined since 2013, decreasing by 6.4% per year, due to the introduction of new treatment strategies, such as targeted therapies for metastatic melanoma and immune checkpoint inhibitors, indicating the importance of developing new diagnostic and treatment modalities.[239] In addition to cancer, skin lesions also pose a significant burden on the healthcare system. The global wound care market is expected to grow from $19.3 billion in 2021 to over $27.8 billion by 2026.[288, 289] Moreover, the global tissue-engineered skin substitutes market is expected to reach $3.9 billion in 2023, with a CAGR of 17.2%.[288] In recent decades, remarkable progress has been made in modeling and developing in vitro-engineered substitutes that mimic human skin[290] for various purposes such as developing in vitro skin models,[291] studying toxicology tests,[292] and creating skin-on-a-chip models for drug testing.[293] 3D bioprinting has further accelerated the relevant science in this field by enabling standardized reproducible fabrication methods using various bioinks[288, 292, 294] or direct cell bioprinting.[133] Although skin-on-a-chip devices[291, 293, 295-297] and 3D-bioprinted skin platforms[298-300] are available in the literature, to our knowledge there is no report on “3D-bioprinted skin-on-a-chip”. Future studies could focus on the development of 3D-bioprinted skin-on-chip to take advantage of both OOC platforms and 3D bioprinting, such as continuous perfusion, precise control of dimensions, and multicell cultures. The fabrication of a bioprinted skin begins with the collection and culturing of cells.[290] Various cells such as keratinocytes, fibroblasts, and melanocytes are cultured and grown after being collected from the human body. The biopolymer matrix is then used to produce the bioink to be used in the bioprinter. Then, the CAD model is prepared depending on the final application. After bioprinting, the fabricated system must undergo different steps in vitro to be ready for further applications such as implantation.
3.10 Multiorgan platforms
The ultimate goal of OOC platforms is to produce multi-OOCs to investigate interactions of different organs more realistically while monitoring possible responses and negative side-effects of administered medicine. However, most of the current OOC platforms are optimized for a specific organ (e.g., particular perfusion rate and circulating medium), complicating the culture of different tissues on a single chip. In this regard, as a proof-of-concept design, a multi-OOC platform was developed using the DLP technology with separate compartments, which enabled the culture of three different types of tissues on a single chip with tissue-specific perfusion rates to study tissue–tissue interactions through interconnective microfluidic channels (1-mm channel radius).[301] To validate the ability of the developed chip to produce different perfusion rates in each compartment independently, 10 and 1 mm3/min flow rates were successfully applied to the first and second wells, respectively, with a laminar flow profile at 0.72 mm/s flow velocity. In addition, the chip was able to generate different shear stresses in each well, recording a shear stress of 0.001 and 0.02 dynes/cm2 on the surfaces of the membrane in chamber one and chamber two, respectively.[301]
4 4D BIOPRINTING—A NEW PARADIGM
The “four-dimensional (4D)-bioprinted OOCs” is a promising field for future research. One of the limitations of 3D bioprinting is considering only the initial state of the cell construct as a static and inanimate structure.[302, 303] 4D bioprinting, as an emerging field of research, introduces “time” into 3D bioprinting by producing bioconstructs that can change shape or functionality after bioprinting in response to an external stimulus (e.g., magnetic or electric field, light, pH, or temperature), cell fusion, or post-printing self-assembly.[302, 304] 4D bioprinting can be accomplished using currently available 3D bioprinters[305] (e.g., nozzle and light based) and smart materials (i.e., materials with inherent ability to reshape themselves in response to external stimuli) or postprint maturation of bioprinted constructs (e.g., matrix deposition, self-organization, or cellular coating).[302, 303] Although 4D printing has been implemented in tissue engineering,[306-309] vascularization,[310] heart stents,[311, 312] nerve repair,[305] and drug delivery,[313] 4D bioprinting is an emerging technology that should be investigated for its potential applications in OOCs. Not only can 4D bioprinting be used to construct more complex 3D structures on chips that are difficult to achieve with typical 3D bioprinters, but it also promotes the biomimetic capability of OOCs by more naturally recapitulating in vivo-like responses and deformations of tissues, when exposed to physiological conditions (e.g., pH changes resulting from drug administration).[71]
In the domain of bioprinting organs and tissues, 4D bioprinting can inaugurate fabrication of bioink-printed microtissues (e.g., cell-laden microgels) that can undertake maturation through the cellular coating, self-organization, and matrix deposition to form functional tissue constructs over time.[314, 315] Furthermore, smart materials and living materials, which can be identified as pioneers enabling 4D bioprinting, are being engineered for a better time and cost efficiency along with resolution improvement and enhanced cell viability.[177-179, 316] While 4D bioprinting has not yet been completely explored for OOC systems, trailblazer similar studies such as the patient-derived organoid-like (PDO) model have been investigated.[317] Enabling drug evaluation in a GBM PDO model, the mentioned 4D bioprinting procedure comprised of 3D printing of a megacassette using a thermo-responsive shape-memory polymer followed by stretching. Next, the shape of the megacassette was transformed to a culture array in a temperature-mediated process. 3D culturing of GBM PDO model was carried out in the 4D array followed by a 4D temperature-mediated shape recovery of the array to a histology megacassette. Not only was this method successful in capturing expression profiles, but also the requirement for manual steps was eliminated.[317]
5 CHALLENGES AND CONCLUDING REMARKS
OOC platforms have superior analytical performance compared with animal models and conventional cell cultures. This superiority results from patient-derived cell cultures with a 3D structure that can more realistically recapitulate the microenvironment in vivo (e.g., cell interactions and responses to stimuli). Continuous perfusion enabled by microfluidic channels promotes cell proliferation and survival while enabling reagent delivery (e.g., drugs) and real-time monitoring of outcomes (e.g., biofluid secretion). 3D bioprinting offers versatility in design and precision in fabrication, ultimately empowering the fabrication of complex 3D organ-mimicking cell constructs on chips. Nevertheless, none of the available 3D bioprinting techniques is the best. Although light-induced methods have high resolutions and can produce complex 3D structures, the high cost, limited material selection, and scalability are still challenging. Also, further studies are needed to determine the conceivable photo-induced damage to cells in the long and short terms. On the other hand, nozzle-based bioprinters are cost effective and available technologies. However, moderate resolution and shear stress (leading to low cell viability) are the major challenges. Future studies can consider developing newer 3D bioprinting methods, or combining current methods to address the challenges with hybrid bioprinters.[71] Although OOC and 3D bioprinting have been extensively explored in the literature, the integration of these two technologies is not yet fully developed. In this review, recent advances, a number of selected end-use applications, and the advantages and disadvantages of various 3D bioprinting methods are presented to highlight the current limitations/challenges of “3D-bioprinted OOCs” and their potential for future applications.
Although primary and immortal cell lines are currently used, further development of cells derived from iPSCs can increase the population diversity required for efficient screening of drugs prior to human clinical trials.[318] Moreover, microscale channels and microchambers allow only a tiny volume of cells to be cultured, resulting in a tiny volume of secreted reagents in the output, which presents a challenge in the precise detection, sampling, and monitoring of results. To address this issue, more precise molecular reporters, nanoscale biosensors, and high-quality imaging devices can be integrated into microfluidic chips to enable accurate real-time monitoring of culture conditions, such as analysis of oxygen, glucose, pH, lactate, fluid pressure, tissue barrier integrity, and cell migration. In addition, the use of parallel microchambers, longer microchannels for culturing, and lower flow rates for perfusion can increase the volume of available reagents. Because of the dominance of viscous forces in microchannels, physical and chemical gradients can be generated by controlling the flow rate and channel dimensions for the noninvasive study of directed cell migration.[39] The use of machine learning (ML) can help researchers determine optimal dimensions and flow rates to achieve desired gradients more cost effectively.[319, 320] Furthermore, ML can assist human experts in quickly and accurately analyzing large acquired data from OOC platforms (e.g., images, videos, and biosensor data).[321]
Perfusion, as one of the main advantages of OOC platforms, is usually performed by embedded micropumps, which increase the cost and complexity of the setups. As a feasible solution, simple and inexpensive gravity-driven and/or auto perfusion systems can be developed by taking advantage of the differential fluid pressure in the reservoirs, which do not require experts to perform cell cultures.[247] In addition, paper-based cell culture platforms can provide a low-cost, biocompatible, and readily available substrate for cell culture with passive capillary-based[322] fluid transfer.[323-326] The porous structure of paper provides a natural 3D support medium for mimicking native cellular microenvironments and creates biointerfaces for cell analysis, including cell-related biochemical analysis (DNA, protein, and small molecules), cell capture/phenotyping, and long-term 3D cell cultures.[323, 327-329] Although cell adhesion and ideal perfusion on paper substrates are still challenging,[323, 330] future studies can focus on overcoming these challenges and 3D bioprinting on paper for OOC applications. Besides, magnetic levitation can play a role in convenient cell separation/isolation/characterization in microfluidic channels at the outlet of chips to enable an analysis of the changes in the concentration of elements in the perfusion medium.[19, 331, 332] An ultimate goal for OOCs is the development of body-on-a-chip platforms that enable patient-specific disease investigation and drug screening considering all organs. However, the media used for perfusion on OOC setups are usually optimized for a specific organ. Therefore, finding a single culture medium (i.e., an alternative for blood) that is compatible with multiple or all organs would be an interesting area of research for the future.
ACKNOWLEDGEMENTS
S. T. acknowledges Tubitak 2232 International Fellowship for Outstanding Researchers Award (118C391), Alexander von Humboldt Research Fellowship for Experienced Researchers, Marie Skłodowska-Curie Individual Fellowship (101003361), and Royal Academy Newton-Katip Çelebi Transforming Systems Through Partnership award (120N019) for financial support of this research. This work was partially supported by Science Academy's Young Scientist Awards Program (BAGEP), Outstanding Young Scientists Awards (GEBİP), and Bilim Kahramanlari Dernegi The Young Scientist Award. N. M. acknowledges Marie Skłodowska-Curie Individual Fellowship (101038093). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. YSZ acknowledges the support by the Brigham Research Institute. Some elements in Figure 1 have been designed using resources from Flaticon.com.
CONFLICT OF INTEREST
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
ETHICS STATEMENT
No human or animal test are conducted in this review article.
Biographies

Sajjad Rahmani Dabbagh is currently a PhD student at Koç University with the focus being on the integration of machine learning and 3D bioprinting for parametric optimization of bioprinting process and defect prediction. His research interests include lab-on-a-chip technologies, point-of-the-care diagnosis, complex fluid mechanics, 3D bioprinting, and artificial intelligence. He graduated from Tabriz University with a bachelor's degree in mechanical engineering in 2016.

Yu Shrike Zhang received a BEng in Biomedical Engineering from Southeast University (2008), an MS in Biomedical Engineering from Washington University in St. Louis (2011), and a PhD in Biomedical Engineering at Georgia Institute of Technology/Emory University (2013). Dr. Zhang is currently an assistant professor at Harvard Medical School and Associate Bioengineer at Brigham and Women's Hospital. Dr. Zhang's research is focused on innovating medical engineering technologies, including bioprinting, organs-on-chips, microfluidics, and bioanalysis, to recreate functional tissues and their biomimetic models toward applications in precision medicine.

Savas Tasoglu received his PhD degree in 2011 from UC Berkeley and held a postdoctoral appointment at Harvard Medical School. His research interests are at the interfaces of complex microfluidics, transport phenomena, and biomaterials with applications in biomedicine and biotechnology. He has received several awards, including a Marie Curie Fellowship, American Heart Association Scientist Development Award, NIH K99/R00 Pathway to Independence Award (declined), Alexander von Humboldt Fellowship for Experienced Researchers, and the Wolff New Venture Competition Award.
Open Research
DATA AVAILABILITY STATEMENT
None.




