Myelin-specific T cells in animals with Japanese macaque encephalomyelitis
Funding Information:
This research was supported by the United States Department of Defense grants [W81XWH-09-1-0276 (LSS, BF, SGK, WDR, and SWW) and W81XWH-17-1-0101 (LSS and SWW)], National Institutes of Health grants that support the ONPRC Flow Cytometry and Japanese Macaque Resource [P51OD011092 (MKA, BF, SGK, LSS, and SWW)] and R24-NS104161 (LSS, BF, SGK, and SWW), The Laura Fund for Multiple Sclerosis Research (DNB and SWW), and the Race to Erase MS Center Without Walls (SWW).
Abstract
Objective
To determine whether animals with Japanese macaque encephalomyelitis (JME), a spontaneous demyelinating disease similar to multiple sclerosis (MS), harbor myelin-specific T cells in their central nervous system (CNS) and periphery.
Methods
Mononuclear cells (MNCs) from CNS lesions, cervical lymph nodes (LNs) and peripheral blood of Japanese macaques (JMs) with JME, and cervical LN and blood MNCs from healthy controls or animals with non-JME conditions were analyzed for the presence of myelin-specific T cells and changes in interleukin 17 (IL-17) and interferon gamma (IFNγ) expression.
Results
Demyelinating JME lesions contained CD4+ T cells and CD8+ T cells specific to myelin oligodendrocyte glycoprotein (MOG), myelin basic protein (MBP), and/or proteolipid protein (PLP). CD8+ T-cell responses were absent in JME peripheral blood, and in age- and sex-matched controls. However, CD4+ Th1 and Th17 responses were detected in JME peripheral blood versus controls. Cervical LN MNCs from eight of nine JME animals had CD3+ T cells specific for MOG, MBP, and PLP that were not detected in controls. Mapping myelin epitopes revealed a heterogeneity in responses among JME animals. Comparison of myelin antigen sequences with those of JM rhadinovirus (JMRV), which is found in JME lesions, identified six viral open reading frames (ORFs) with similarities to myelin antigen sequences. Overlapping peptides to these JMRV ORFs did not induce IFNγ responses.
Interpretations
JME possesses an immune-mediated component that involves both CD4+ and CD8+ T cells specific for myelin antigens. JME may shed new light on inflammatory demyelinating disease pathogenesis linked to gamma-herpesvirus infection.
Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating disease (IDD) of the central nervous system (CNS) that leads to the destruction of the myelin sheaths that surround the axons of CNS neurons. Histopathological examination of demyelinating lesions identifies multiple cellular components and products of the immune system, including activated macrophages, CD4+ and CD8+ T cells, B cells, antibodies, complement component C3d, and proinflammatory cytokines.1 Importantly, extensive population-based studies have reported that genetics and environmental factors are involved in MS, and epidemiology studies have proposed that a human herpesvirus infection could induce IDD in MS. Specifically, Epstein–Barr virus (EBV) or human herpesvirus 6 (HHV-6) are postulated to be potential viral triggers. Exactly how virus infection, the immune components, genetics, and other environmental factors coalesce to induce and facilitate the inflammatory condition that ultimately leads to demyelination and axonal injury is poorly understood.
Animal models that mimic IDD are widely recognized for improving our understanding of how the immune system attacks myelin. There are two frequently utilized animal models that are studied. The most extensively utilized animal model to study MS is experimental autoimmune encephalomyelitis (EAE), which can be produced in a variety of species, including mice, rats, and non-human primates (NHP), involves immunizing animals with myelin proteins or peptides in Freund’s complete adjuvant, which results in T-cell responses to myelin, leading to focal inflammatory lesions within the CNS and ultimately paralysis. EAE recapitulates the T cell-mediated aspects of MS, as studies find Th1 and Th17 cells are necessary for the induction of EAE.2 However, while EAE studies have yielded useful insight into several facets of MS pathogenesis, this model has well-recognized limitations. First, the immunization regime artificially induces the disease, while MS occurs as a spontaneous disease. And second, EAE is studied primarily in inbred mouse strains and this is in large contrast to MS, which occurs in a heterogeneous population with highly variable genetic diversity. More recently, however, others have shown that induction of EAE with recombinant myelin oligodendrocyte glycoprotein (MOG) encoding the extracellular domain (aa 1-170) or MOG encephalitic peptide (aa 34-56) emulsified in incomplete Freund’s adjuvant in cynomolgus macaques (Macaca fascicularis) can lead to atypical EAE disease that displays heterogeneity in clinical course, including less progressive disease accompanied with periods of remission.3 Additionally, atypical EAE disease in macaques includes comparable MRI results, demyelination, and MOG-specific T-cell proliferation resulting in Th1 and Th17 responses.4 Like EAE in marmosets, the macaque model involves outbred animals rather than inbred rodents, which more closely resembles the human population.
The second most common animal model is the Theiler’s murine encephalomyelitis virus (TMEV) infection of SJL/J mice. TMEV is a picornavirus that was isolated from the CNS of mice with flaccid hind limb paralysis.5 The model was developed in 1975 and involves intracerebral injection of TMEV (Daniel strain) into susceptible mice, resulting in persistent virus infection and a chronic progressive biphasic disease, referred to as TMEV-inflammatory demyelinating disease (TMEV-IDD).6 Importantly, TMEV-IDD provides a valuable animal model to elucidate how virus infection can lead to CD8+ T-cell responses to myelin epitopes. Four different methods for how virus infection can trigger immune-mediated demyelination have been postulated and these include: epitope spread, molecular mimicry, dual T-cell receptor, and virus-mediated programing of antigen-presenting cells (reviewed in 7). Understanding whether these methods or undefined mechanisms are associated with the development of MS requires an animal model that closely approximates disease in humans.
Interestingly, a hybrid virus/EAE model is currently being investigated to elucidate how EBV infection plays a role in MS induction. One approach utilizes the EAE/marmoset model and EBV or related primate lymphocryptovirus infection of B cells. Results from these studies suggest that virus-infected B cells process pathogenic MOG epitope for presentation to reactive CD8+ T cells.8 A second research team reports that latent murine herpesvirus 68 (MHV-68), a rodent gamma-herpesvirus related to EBV and Kaposi’s sarcoma-associated herpesvirus (KSHV), enhances autoimmunity leading to severe EAE disease pathology by upregulating CD40 expression on uninfected antigen-presenting cells during latent MHV-68 infection and increasing both CD4+ and CD8+ effector T-cell activation, while reducing T regulatory cells during EAE.9, 10 Both approaches pose novel mechanisms of action as to how gamma-herpesvirus infection can exacerbate immune-mediated demyelination.
New animal models that mimic the pathophysiology of MS will greatly aid in understanding how MS is initiated and will accelerate the development of novel MS therapies. One model that could facilitate this is the spontaneous Japanese macaque encephalomyelitis (JME) model that appeared in the Japanese macaque (JM) colony at the Oregon National Primate Research Center (ONPRC). JME displays clinical, histopathological, and immunopathological parallels with MS and is linked to infection by a novel gamma-herpesvirus, JM rhadinovirus (JMRV).11-13 However, the acute nature of JME has led others to consider JME as a model of acute demyelinating encephalomyelitis (ADEM), especially since disease is linked to a virus infection or that JME is a distinct IDD.14 To further elucidate the immunopathogenesis of JME, we investigated the nature of the T-cell response using intracellular cytokine staining (ICS) coupled with flow cytometry (FCM) and ELISpot assays to myelin antigens. With these immunological assays, we found that JME animals possess myelin-reactive CD4+ and CD8+ T cells that target myelin oligodendrocytes glycoprotein (MOG), myelin basic protein (MBP), and proteolipid protein (PLP), analogous to MS pathophysiology.15-18 Moreover, by mapping overlapping epitopes, we found that two JM possess homologous antigenic epitopes detected in MS patients.18, 19 These studies support JME as a spontaneous NHP model for IDD, and demonstrate that more investigation is warranted to define the etiology of the disease, which could apply to MS.
Subjects and Methods
Animals
All animal protocols and procedures were reviewed and approved by the ONPRC Institutional Animal Care and Use Committee. The ONPRC is an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International accredited research facility and conforms to the National Institutes of Health (NIH) guidelines on the ethical use of animals in research. JMs exhibiting neurological dysfunction and symptoms associated with JME were brought in for physical examination and provided supportive care, and then scanned by MRI on a 3T Siemens TIM Trio MR instrument and humanely euthanized for tissue collection as previously described.11 Blood was immediately processed for serum and plasma collection and peripheral blood mononuclear cell (PBMC) isolation. Lymph nodes (cervical, axillary, and inguinal) were collected and processed for mononuclear cell (MNC) isolation. The CSF, plasma, and serum were archived at −80°C, while PBMCs and MNCs were cryopreserved. Animals were subsequently perfused with sterile PBS through carotid cannulation, and lesion areas that were detected by MRI were collected from affected areas after the PBS perfusion step in order to obtain fresh brain tissue for inflammatory cell analysis. A portion of each lesion was placed in RPMI media supplemented with 10% fetal bovine serum and processed as described to isolate CNS-infiltrating mononuclear cells (CNS-MNCs).12 In these cases, the remainder of the brain was immersion fixed in 4% neutral buffered paraformaldehyde solution for histopathological analysis. For comparisons to JME cases, cervical lymph nodes (CV LNs) were collected from JM scheduled for humane euthanasia and these tissues were processed as above for MNCs and cryopreserved.
Histopathological examination
Fixed tissue was processed for paraffin embedding, sectioned (0.5 mmol/L), and stained with Luxol fast blue (LFB) and hematoxylin and eosin (H&E) to visualize demyelinating regions and infiltrating inflammatory cells. Sections containing lesions were treated essentially as described previously for evidence of inflammatory demyelination.12
CNS-mononuclear cell (MNC) stimulation and flow cytometry analysis
Freshly purified CNS-MNCs were isolated, stimulated with a cocktail of phorbol-12-myristate-13 acetate (PMA) and Ionomycin that included Brefeldin A (Leukocyte Activation Cocktail with BD GolgiPlug, BD Biosciences, San Jose, CA, USA) for 6 h, and analyzed as described previously.12
Myelin-specific CD4+ and CD8+ T-cell responses were measured in cryopreserved CNS-MNC and PBMC preparations (2 × 106 cells) from JME animals and age- and gender-matched healthy controls (HC) by flow cytometric ICS (FCM-ICS).20 Thawed and warmed CNS-MNC and PBMCs were incubated with peptide pools comprised of sequential 15-mer peptides overlapping by 11 amino acids (aa) representing macaque myelin oligodendrocytes glycoprotein (mMOG, NCBI: NP_001181792.2), macaque myelin basic protein (mMBP, GenBank: EHH29376.1), and macaque proteolipid protein (mPLP, GenBank: AFJ71086.1) (Sigma-Aldrich, St. Louis, MO, USA) with co-stimulatory CD28 and CD49d monoclonal antibodies (0.5 µg/mL, BD Biosciences) for 1 h, followed by addition of Brefeldin A for an additional 8 h.21 Staphylococcal enterotoxin B (SEB, Toxic Tech) was used as a positive control and DMSO only as negative control. Individual peptides were present at 2 µg/mL per assay. After antigen stimulation, the viability of cells was assessed using LIVE/DEAD Aqua fixable stain (Invitrogen). The following antibodies were used prior to fixation: CD4 (L200, BD), CD8-α (SK1, Biolegend), and CD45 (H130, Invitrogen). Cells were then fixed and permeabilized with BD Lyse (BD) and Tween-20 (Sigma) and stained with the following antibodies specific for: CD3 (SP34-2, BD), CD69 (FN50, BD), IFN-γ (B27, BD), TNF-α (B27, eBioscience), and IL-17A (eBio64CAP17, eBioscience). Positive responses were determined by first removing CD45-negative cell populations and then selecting for CD3+ cells. This CD3+ cell population was further gated for lymphocytes, followed by gating for singlets, and then live cell populations. Within this population, gates were drawn for CD4+ or CD8+ single positive cells expressing CD69. Next, cytokine-expressing cells (IFNγ+ or IL-17+) were gated within these T-cell populations. Finally, background responses from negative controls (co-stimulatory antibodies and DMSO only) were subtracted from this value. All FCM data were acquired on LSR II (BD) and analyzed using Flow Jo (Tree Star).
ELISPOT assay
Frozen MNC suspensions from CV LNs were thawed, washed, and rested at least 1 h at 37°C with 5% CO2. The rested MNCs were used in IFNγ-ELISPOT assays for the detection of IFNγ-secreting cells as previously described.22 Approximately 1 × 105 cells were added to each well along with a 1 µmol/L concentration of a peptide pool and incubated overnight at 37°C with 5% CO2. Initially, the MNCs were screened with mMOG, mMBP, and mPLP pools of 10 peptides. These pools comprised the entire protein for each myelin-associated antigen in the screen. SEB and DMSO and no peptide were used as positive and negative controls, respectively. Each sample was tested in duplicate. Positive results from the initial peptide pool screen were then tested with a 1 µmol/L concentration of the individual myelin-associated 15-mer peptides. Plates were processed on day 2 according to the manufacturer’s instructions (Monkey IFNγ ELISpot Plus, MabTech 3420M-4APT-10). The plates were read using an AID ELISpot reader and software, version 4.0 (Strasburg, Germany). Responses were considered positive if the mean number of spot-forming cells (SFC) of duplicate sample wells exceeded the background plus two standard deviations. Responses of less than 5 SFC per 100,000 CV LN MNC were considered negative. Positive responses were determined using a one-tailed t test and an alpha level of 0.05, with a null hypothesis that the background level would be greater than or equal to the treatment level. If statistically determined to be positive, then the values were reported as the average of the test wells minus the average of the highest negative control wells.
Myelin epitope analysis
BLAST search was performed with experimentally identified MOG, MBP, and PLP peptide epitopes with all rhadinovirus proteins, including those potential proteins encoded by JMRV to identify similar peptide sequences. Analysis was performed using the Virus Pathogen Resource Website (www.vipbrc.org) and Analyze & Visualize program for peptide sequence comparison.
Measurement of JMRV-specific antibody titers
Anti-JMRV IgG antibody titers, plasma, or serum collected at necropsy or from scheduled physical examinations were measured using a standard enzyme-linked immunosorbent assay (ELISA).23 For these experiments, serial threefold dilutions of plasma/serum were incubated in duplicates on JMRV virus lysate-coated ELISA plates for 1 h prior to washing, staining with detection reagents (HRP-anti-IgG), and addition of chromogen substrate to allow for detection and quantitation of bound antibody molecules. Log–log transformation of the linear portion of the curve was then performed, and 0.15 OD units were used as the cut-off point to calculate end-point titers. Each plate included a positive control sample used to normalize the ELISA titers between assays, and a negative control sample to ensure the specificity of the assay conditions.
Statistical analysis
T-cell phenotypes and responses to MOG, MBP, and PLP overlapping peptide pools in the PBMCs from JME and HC were analyzed for statistical analysis using GraphPad Prizm (GraphPad Software, La Jolla, CA, USA), and significant differences in the means were determined by an unpaired t test, with P values of ≤0.05 considered significant.
Results
Nine JMs that presented with clinical signs consistent with JME were evaluated by MRI to determine if CNS lesions were present. All animals had two or more T2-weighted lesions, with the cerebellum being the most affected site (eight animals), followed by the cervical spinal cord (seven animals) and brain stem (six animals) (Table 1). Histopathological analysis of the CNS lesions confirmed IDD with seven of nine lesions staining positive for CD163+ cells engulfing MBP, and all lesions staining positive for IL-17 in either CD3+ cells, Olig-2+ cells, or GFAP+ cells.
| Animal ID | MRI lesion location | Age (years/days) | Gender | Histopathology | IL-17+ staining | MHC class I | Anti-JMRV IgG titer |
|---|---|---|---|---|---|---|---|
| 24041 | CBM, BS | 13y 154d | M | CD163+/MBP+ | CD3+/Olig2+ | -A04,-A01,-B*050,-B*078,-B*011 | 3,199 |
| 29756a | CBM | 5y 289d | M | CD163+/MBP+ | CD3+/GFAP+ | -A01,-A04,-B*017,-B*057,-B*069 | 664 |
| 31875 | CBM | 4y 297d | F | CD163+/MBP+ | CD3+/Olig2+/GFAP+ | -A01,-A04,-B*050,-B*078,-B*011 | 2,345 |
| 31869a | CBM, BS | 3y 156d | F | CD163+ | CD3+/Olig2+ | -A04,-A01,-B*029,-B*017,-B*050,-B*078 | 2,415 |
| 34361 | C-SC | 1y 107d | F | CD163+/MBP+ | Olig2+/GFAP+ | -A01,-B*050,-B*078,-B*060 | 987 |
| 34027a | CBM, BS, BF | 0y 323d | F | CD163+/MBP+ | CD3+/Olig2+ | -A02,-A01,-A04,-B*017 | 1,617 |
| 34421a | CBM | 0y 323d | F | CD163+/MBP+ | CD3+/Olig2+/GFAP+ | -A01,-A04,-B*050,-B*078,-B*060,-B*082 | 1,036 |
| 34478a | CBM | 0y 215d | F | CD163+ | CD3+/Olig2+/GFAP+ | -A01,-B*060,-B*036,-B*029,-B*050,-B*078 | 0 |
| 35604 | CBM | 0y 149d | F | CD163+/MBP+ | CD3+/Olig2+ | -A04,-A02,-B*050,-B*078,-B*036 | 1,031 |
| 19895b | NA | 14y 132 | M | NA | NA | -A01,-A04,-B*050,-B*078,-B*162 | 1,861 |
| 29737b | NA | 5y 296d | M | NA | NA | -A04,-A06,-B*050,-B*078 | 252 |
| 28581b | NA | 5y 331d | F | NA | NA | -A04,-B*050,-B*078 | 4,377 |
| 30217b | NA | 5y 306d | F | NA | NA | -A04,-A06,-B*050,-B*078,-B*011 | 3,504 |
| 34046b | NA | 1y 135d | F | NA | NA | -A04,-A02,-B*050,-B*078,-B*101 | 828 |
| 34040b | NA | 1y 135d | F | NA | NA | -A01,-B*050,-B*078,-B*060,-B*070 | 1,480 |
| 29202b | NA | 1y 114d | F | NA | NA | -A02,-A04,-A05,-B*101,-B*070 | 2,512 |
| 32930b | NA | 1y 114d | F | NA | NA | -A01,-A04,-B*050,-B*078-B*162 | 1,102 |
| 30765b | NA | 1y 105d | F | NA | NA | -A05,-A06,-A01,-B*050,-B*078,-B*070,-B*011 | 2,564 |
| 34038b | NA | 1y 73d | F | NA | NA | -A02,-A04,-B*050,-B*078,-B*101,-B*051 | 2,303 |
| 34358b | NA | 0y 308d | F | NA | NA | -A01,-A02,-B*050,-B*078,-B*060 | 1,861 |
| 17807c | NA | 22y 268d | F | NA | NA | ND | 2,731 |
| 21247c | NA | 16y 29d | F | NA | NA | ND | 1,450 |
| 26176c | NA | 10y 351d | F | NA | NA | ND | 5,980 |
| 33720c | NA | 5y 326d | F | NA | NA | ND | 1,809 |
| 34037c | NA | 5y 293d | F | NA | NA | NA | 1,049 |
| 34364c | NA | 5y 44d | F | NA | NA | ND | 5,697 |
| 34954c | NA | 4y 290d | M | NA | NA | ND | 1,883 |
| 34978c | NA | 4y 343d | M | NA | NA | ND | 922 |
| 34980c | NA | 4y 337d | M | NA | NA | ND | 1,662 |
| 36978c | NA | 2y 359d | F | NA | NA | ND | 1,805 |
| 36993c | NA | 3y 11d | F | NA | NA | ND | 4,626 |
| 38114c | NA | 1y 360d | F | NA | NA | NA | 933 |
- CBM, Cerebellum; BS, Brainstem; C-SC, Cervical Spinal Cord; BF, Basal Forebrain.
- CD163+/MBP+: CD163+ cells engulfing MBP and CD163+: CD163+ cells infiltrating lesion.
- NA, Not applicable; ND, Not determined.
- a CNS MNCs were isolated and utilized in Figure 1A, B, and C.
- b HC: Healthy control PBMCs utilized in Figure 1C.
- c Control JM CV LN MNC utilized in Figure 2B or in experiment Figure 4.
As five of the JME animals harbored lesions in both hemispheres of the cerebellum, fresh lesion tissue from one hemisphere was collected and processed for infiltrating CNS-MNCs. A fraction of the CNS-MNC infiltrates were first stimulated with PMA and ionomycin and analyzed by ICS for IL-17, IFNγ, and immunophenotyped for CD3+, CD4+, or CD8+ T cells by FCM.12 All five animals had T-cell infiltrates with Th1, Th17, CTL, or Tc17 phenotypes similar to our previous report.
We next interrogated the CNS-infiltrating T cells for myelin-specific responses utilizing serial 15-mer peptides overlapping by 11 aa representing macaque MOG, MBP, and PLP, followed by cell surface phenotyping for CD4, CD8, and CD45, and ICS for CD3, CD69, IFNγ, IL-17A, and TNFα. As controls, DMSO (negative control), PMA/ionomycin (positive control), and SEB (positive control) were included in our analysis. A representative FCM scheme and gating strategy are provided for animal 31869, which starts by removing contaminating debris from the CNS lesion by selecting for the CD45+ cell population, followed by gating on the CD3+ cell population, and subsequently for single lymphocytes. This CD45+CD3+ lymphocyte population is then separated into CD4+ and CD8+ populations and then for cytokine staining (Fig. 1A). By this analysis, two JMs possessed CTL to myelin components. JM 34478 possessed CTL to MBP (6.15%) and MOG (3.29%), and JM 31869 had CTL response to MBP (2.06%). Three JMs displayed myelin-specific Th1 responses, with JM 34027 having the highest frequencies to MOG (7.46%), MBP (4.17%), and PLP (9.3%), followed by JM 31869 with Th1 responses to all three myelin antigens (MOG, 1.28%; MBP 3.36%; and PLP 1.02%), and JM 34421 possessing Th1 response to MOG (0.11%). Three JMs (29756, 34027, and 31869) possessed Th17 cells to at least two myelin antigens, with JM 34478 and 34421 possessing no Th17 responses. JM 31869 was the only JM to possess Tc17 responses to any myelin antigens (PLP, 1.39%) (Fig. 1B).
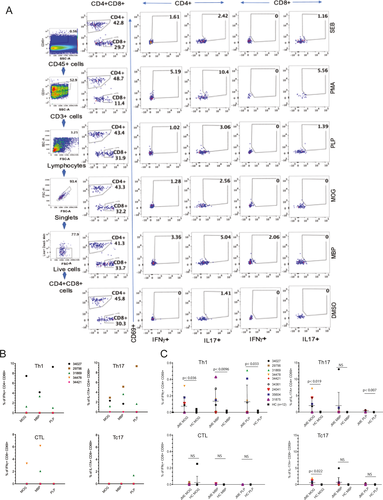
To determine if T-cell responses in the CNS are represented in the PBMCs, we analyzed PBMCs from the nine JME animals and 11 age- and sex-matched healthy controls (HC, n = 11). In humans, HC have been reported to possess myelin-specific T cells in peripheral blood.24, 25 Interestingly, seven of nine JME animals displayed Th1 responses in the blood to MOG, MBP, or PLP, whereas five of nine JME animals possessed Th17 cells in the blood. Most notable, JM 29756 and 31869 did not possess Th17 responses to MOG, MBP, or PLP in the blood, despite having Th17 responses in the CNS. CTL and Tc17 responses in the blood were less representative, with only 29756, 24041, and 31875 displaying CTL responses to MOG or MBP, and JM 24041, 31875, 34027, 34361, 34478, and 35604 possessing Tc17 responses to MOG, MBP, or PLP. Overall, JME animals possessed detectable Th1 and Th17 responses to MOG, MBP, and PLP in PBMCs that were statistically significant when compared to the HC (Fig. 1C).
We next determined whether we could identify myelin-specific T-cell epitopes in JME animals. To accomplish this, we utilized MNCs from disrupted CV LN, as the CNS-MNCs were exhausted for the experiments in Figure 1A and B, and the PBMCs did not represent the CNS T-cell responses to myelin antigens. We were confident with this approach, as others have reported that CV LN MNCs from animals with EAE or patients with MS harbor antigen-presenting cells positive for myelin adjacent to T cells, implying effective antigen presentation.26, 27 Thus, cryopreserved preparations of CV LN MNCs from the nine JME animals and four control JM were evaluated by IFNγ-ELISpot assays for T-cell responses. Approximately 1 × 105 thawed and rested CV LN MNCs were pulsed with peptide pools of MOG (pools A-F), MBP (pools A-G), or PLP (pools A-F) and then analyzed by ELISpot assay for IFNγ production. Each pool contained 10 or fewer sequential peptides, to ensure appropriate concentrations for assays. Results from IFNγ-ELISpot assays from JM 34027, 34361, 34421, and 35604 are shown with myelin peptide pools that yielded spots forming cells over background (SFC-OB), and include positive (SEB) and negative controls (media and DMSO) (Fig. 2A). By this analysis, JM 24041, 29756, 31869, 34027, 34361, 34421, 34478, and 35604 had SFC-OB responses with varying intensities, whereas JM 31875 and the four controls exhibited no responses (Fig. 2B).
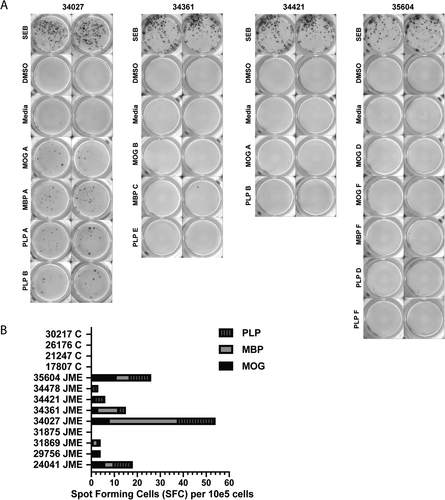
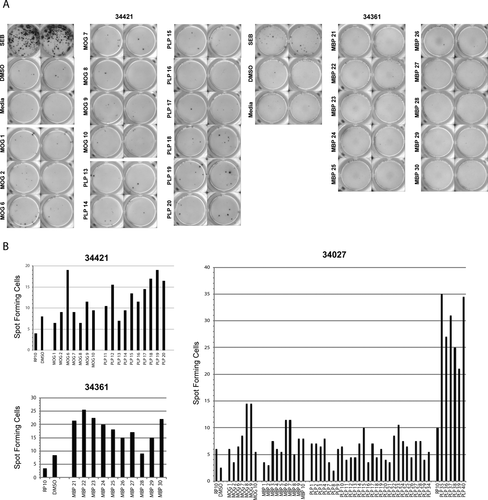
To identify and define the myelin-specific epitopes, individual peptides from the reactive pools were analyzed with CV LN MNC from the eight JME animals. IFNγ-ELISpot assays for CV LN MNCs from 34027, 34361, and 34421 with individual peptides from the reactive pools yielded SFC-OB, whereas MNCs from 24042, 29756, 31876, 34478, and 35604 with individual peptides from specific reactive pools did not (data not shown). ELISpot assays with CV LN MNC from 34421 and 34361 are shown in Figure 3A. As MNCs from 34421 were reactive to MOG peptide pool A and PLP peptide pool B, individual peptides from these pools were plated and SFC-OB was observed with MOG peptide 6 (10 SFC-OB), and PLP peptides 12, and 15-20 (1-9 SFC-OB), whereas MNCs from 34361 were reactive to MBP peptides 21-27 and 29-30 (5-15 SFC-OB). CV LN MNCs from 34027 were reactive to six peptide pools (MOG A, MBP A, and PLP A, B, C, and D) from our initial screen. MNCs were most reactive to PLP peptides 35-40 (13-27 SFC-OB), followed by MOG peptides 8 and 9 (6 SFC-OB), and MBP peptides 6 and 7 (3 SFC-OB) (Fig. 3B).
We next aligned the peptide sequences from those peptides yielding SFC in MOG (peptides 6–9), MBP (6–7 and 21–24), and PLP (11–13, 17–20, 29–30, and 35–40) to define T cell-specific epitopes recognized by animals 34027, 34361, and 34421 (Table 2). Alignments of the MBP peptides capable of inducing SFC-OB did not coincide with those reported to induce T-cell responses in MS patients.28 However, when the MOG and PLP peptides were aligned, we identified homologous T cell-specific epitopes in both MOG and PLP that were identified in human MS (Fig. 4A). Specifically, the MOG peptide sequence (LLQVSSSYAGQF-RVIGPRQPIRALVGD) is homologous with a human MOG peptide sequence that has been reported to be recognized in human MS.29 While only one MOG peptide sequence was identified, we found two of the three macaque PLP peptide sequence regions that are immunodominant in MS patients (KLIETYFSKNY and IYFNTWTTCQSIAFPSKTSA).16, 19, 30 These data demonstrate that animals with JME can harbor myelin-specific T-cell responses to myelin epitopes that have been identified in some MS patients.
| Peptidea | Sequenceb | Spot-forming cells over background (SFC-OB) |
|---|---|---|
| MOG6 | LLQVSSSYAGQFRVI | 11 |
| MOG7 | SSSYAGQFRVIGPRQ | 2 |
| MOG8 | AGQFRVIGPRQPIRA | 8 |
| MOG9 | RVIGPRQPIRALVGD | 8 |
| consensus c | LLQVSSSYAGQFRVIGPRQPIRALVGD | |
| MBP6 | TNRGESEKKTNLGEL | 6 |
| MBP7 | ESEKKTNLGELSRTT | 6 |
| consensus | RGESEKKTNLGELSR | |
| MBP21 | HPADPGSRPHLIRLF | 13 |
| MBP22 | PGSRPHLIRLFSRDA | 17 |
| MBP23 | PHLIRLFSRDAPGRE | 14 |
| consensus | GSRPHLIRLFSRD | |
| MBP28 | DRPSESDELQTIQED | 0 |
| MBP29 | ESDELQTIQEDSAAT | 6 |
| MBP30 | LQTIQEDSAATSESL | 13 |
| consensus | LQTIQEDSAATSESL | |
| PLP11 | TGTEKLIETYFSKNY | 3 |
| PLP12 | KLIETYFSKNYQDYE | 8 |
| PLP13 | TYFSKNYQDYEYLIN | 0 |
| consensus | KLIETYFSKNYQDYE | |
| PLP17 | IHAFQYVIYGTASFF | 6 |
| PLP18 | QYVIYGTASFFFLYG | 9 |
| PLP19 | YGTASFFFLYGALLL | 11 |
| PLP20 | SFFFLYGALLLAEGF | 8 |
| consensus | YGTASFFFLYGALLL | |
| PLP35 | VPVYIYFNTWTTCQS | 25 |
| PLP36 | IYFNTWTTCQSIAFP | 17 |
| PLP37 | TWTTCQSIAFPSKTS | 21 |
| PLP38 | CQSIAFPSKTSASIG | 15 |
| PLP39 | AFPSKTSASIGSLCA | 11 |
| PLP40 | KTSASIGSLCADARM | 24 |
| consensus | Y IYFNTWTTCQSIAFPSKTSASIGSLCADARM |
- a Primary amino acid (aa) sequence of 15-mers overlapping by 11 aa.
- b Peptide sequences inducting spot-forming cells (SFC) from animals 34421, 34361, and 34027 were aligned to identify T-cell epitopes.
- c Consensus sequence of myelin epitope defined by ELISpot analysis
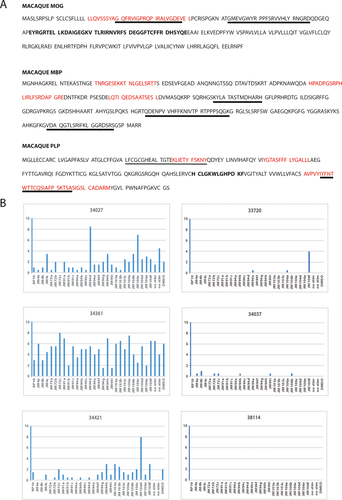
As we had earlier isolated and characterized a novel gamma-2 herpesvirus [JMRV] from a JME lesion, we utilized in silico analysis to determine if protein sequences derived from JMRV and all known rhadinoviruses, including rodent rhadinovirus MHV-68, harbored aa sequence similarity/identity with the MOG, MBP, and PLP responsive peptides. BLAST search using the myelin-specific peptide sequences in Figure 4A did not reveal homologous sequences, whereas short peptide sequences of MOG found one region within JMRV ORF 72 (PIRALVG), the three regions of MBP yielded sequence homology with two regions within JMRV ORF 94 (GELSR and HLTRLF), one with JMRV ORF 103 (RGVEESEK), and two with JMRV ORF 106 (IRLSSR). Two of the sequences in PLP yielded homology with JMRV ORF 94 (TASFF), JMRV ORF 91 (FFLSGA), and JMRV ORF 4 (NTTTTC). To test if these JMRV peptides serve as molecular mimics of MOG, MBP, and PLP epitopes, we had three 15-mer overlapping by 11 aa peptides corresponding to the JMRV ORF sequence synthesized for ELISpot assays. The aa sequence of each peptide is shown along with the potential mimic sequence from the myelin antigen highlighted in red with mismatches in black (Table 3). Using these overlapping peptides, we did not detect peptides that yielded positive SFC-OB with CV LN MNCs from JME animals 34027, 34361, and 34421 or from 12 JMRV seropositive age- and gender-matched control JMs (Fig. 4B). These data imply that molecular mimicry between myelin epitopes and JMRV is not a mechanism for immune-mediated demyelination in JME.
| Myelina/JMRVb ORF | Amino acid sequence |
|---|---|
| PLP | AVPVYIYFNTWTTCQSIAFPSKTS |
| JMRV-4 | TITFTCNKDFSLIGNTTTTCMTNGTWSSPV |
| JMRV-4a | NKDFSLIGNTTTTCM |
| JMRV-4b | SLIGNTTTTCMTNGT |
| JMRV-4c | NTTTTCMTNGTWSSP |
| MOG | SYAGQFRVIGPRQPIRALVGDEVELPCRIS |
| JMRV-72 | AEHAKLCQLLNTAPLKALVGSTCNDMYKDI |
| JMRV-72a | LCQLLNTAPLKALVG |
| JMRV-72b | LNTAPLKALVGSTCN |
| JMRV-72c | PLKALVGSTCNDMYK |
| PLP | QYVIYGTASFFFLYGALLLAEGFYTTGAVR |
| JMRV-91 | LRIIRAPQCAFFLSGAPTDEVYYHTGLIDQ |
| JMRV-91a | IIRAPQCAFFLSGA |
| JMRV-91b | PQCAFFLSGAPTDEV |
| JMRV-91c | FFLSGAPTDEVYYHT |
| PLP | IHAFQYVIYGTASFFFLYGALLLAEGF |
| JMRV-94 | TSGDMAEVITDILTGTQATASFFCVLHDRGNVPINTPHAV |
| JMRV-94a | ITDILTGTQATASFFC |
| JMRV-94b | TGTQATASFFCVLHD |
| JMRV-94c | ATASFFCVLHDRGNV |
| MBP | HPADPGSRPHLIRLFSRDAPGRE |
| JMRV-94 | GTFYFFQSYTCTAETLVHLTRLFISSQGQSLVTVNTYDEL |
| JMRV-94d | TCTAETLVHLTRLFI |
| JMRV-94e | ETLVHLTRLFISSQG |
| JMRV-94f | HLTRLFISSQGQSLV |
| MBP | TNRGESEKKTNLGELSRTTSEDSEVF |
| JMRV-94 | VRVPHCYKVGPGGELSRLLKIIICHPEESD |
| JMRV-94g | PHCYKVGPGGELSRL |
| JMRV-94h | KVGPGGELSRLLKII |
| JMRV-94i | GGELSRLLKIIICHP |
| MBP | TNRGESEKKTNLGELSRTT |
| JMRV-103 | IPWNHDRGSRGVEESEKNIFIDYCRSGIL |
| JMRV-103a | WNHDRGSRGVEESEK |
| JMRV-103b | RGSRGVEESEKNIFI |
| JMRV-103c | GVEESEKNIFIDYCR |
| MBP | PSESDELQTIQEDSAATSESLDVMASQKR |
| JMRV-106 | PGLFWADEHKTRLVLAATSPSLPNYDYQRD |
| JMRV-106a | EHKTRLVLAATSPSL |
| JMRV-106b | RLVLAATSPSLPNYD |
| JMRV-106c | AATSPSLPNYDYQRD |
| MBP | PGSRPHLIRLFSRDAPGRE |
| JMRV-106 | AELSTATGQGIRLSSRPTNKSGHHVCVLDG |
| JMRV-106a | LSTATGQGIRLSSRP |
| JMRV-106b | TGQGIRLSSRPTNKS |
| JMRV-106c | IRLSSRPTNKSGHHV |
- a Macaque myelin antigens; phospholipid protein (PLP), myelin oligo-glycoprotein (MOG), myelin basic protein (MBP) with potential mimic sequence in red.
- b Japanese macaque rhadinovirus (JMRV) open reading frame (ORF) with aa sequence homology with myelin antigen. JMRV-4, JMRV Complement regulatory protein; JMRV-72, capsid protein; JMRV-91; viral dUTPase; JMRV-94, Helicase/primase; JMRV-103, viral interferon regulatory factor R8; JMRV-106, viral interferon regulatory factor R10.
Discussion
In this report, we undertook an immunological-based study to determine whether the CNS-infiltrating T cells from five JME animals exhibit specificity for myelin antigens. We stimulated CNS-MNCs with peptide pools representing macaque MOG, MBP, and PLP, followed by ICS and FCM. All five JMs possessed myelin-specific T cells in the affected CNS tissues, with CD4+ and CD8+ T-cell responses detected for the three myelin antigens. Interestingly, not all of the IFN-γ− or IL-17-producing T cells were specific for myelin antigens, implying only a subset of the Th1 and Th17 are associated with disease, much like what has been reported in human MS.31 Importantly, this suggests that there could be other T-cell epitopes associated with JME that contribute to CNS immunopathology.
The ability to map the antigenic T-cell epitopes associated with JME was one of our goals for this study if there were myelin-specific T-cell responses. Unfortunately, the recovery of MNCs infiltrating the CNS lesions did not yield sufficient number for interrogation. And, since the T-cell responses we detected in the CNS were not fully represented in the blood, we utilized MNCs from the draining cervical LNs, which have been reported to harbor professional antigen-presenting cells staining positive for myelin juxtaposed to T cells in macaques with EAE and in MS patients.26, 27 Using IFNγ ELISpot assays, we identified one MOG-, three MBP-, and three PLP-specific aa sequence regions that CV LN MNCs from JME animals target. Further analysis revealed that one MOG-specific sequence (aa 30-49) and two PLP-specific sequences (aa 44-55 and 176-195) are homologous T-cell targets in MS. Interestingly, one PLP-specific sequence, aa 176-195, was published to be targeted by T cells from MS patients with HLA-DQB1*601 loci of the HLA-DR15 haplotype.30
As CD4+ and CD8+ T cells play a prominent role in MS, we sought to determine the MHC class I and II alleles, as multiple MHC I and II alleles have been reported to increase the risk of MS.31-34 To define if conserved MHC 1 alleles increase JME risk, we used PCR primers for Rhesus macaque MHC class I alleles that can detect JM MHC class I alleles to determine whether specific MHC class I alleles are overrepresented in JME animals compared to those in HC from the JM colony. We found that MHC class I alleles did not differ significantly between JME animals and HC, with major MHC class I alleles equally represented in both JME and HC (Table 1). However, we did identify specific alleles (B*017, B*029, B*036, and B*082) in six of nine JME animals that were not represented in HC, and alleles in the HC (A05, A06, B*051, B*070, B*101, and B*162) that were not present in JME animals. Efforts to define MHC-II alleles within the JM colony are underway and could shed additional light as to how JME tracks with specific JM matrilines.11
Our data also suggest that molecular mimicry with JMRV, a gamma-herpesvirus we found in JME lesions, is not likely a potential mechanism for induction of JME, as JMRV peptides harboring near identical aa sequences as the reactive myelin peptides did not yield IFNγ+ ELISpots. However, it should be noted that only five aa in the JMRV peptides were homologous with the myelin antigens. Thus, this does not necessarily prove that JMRV is or is not associated with induction or development of JME. The results only demonstrate that T-cell responses to myelin-specific peptides in this group of JME animals did not react to JMRV peptides that possessed the homologous five aa sequence.
Thus far, our cumulative data demonstrate that JME possesses features that are similar to MS, including comparable MRI results, inflammatory demyelinating lesions that resemble MS lesions, positive CSF findings, CNS-infiltrating T cells exhibiting Th1 and Th17 phenotypes, and ex vivo evidence of CNS-infiltrating T cells targeting myelin-specific antigens. So, how does JME compare to other NHP models of MS? JME shares some similarity with atypical EAE in macaques, as lesion development and clinical course differ between animals, and both yield T cell-specific responses to the same MOG sequences. As an NHP model for immune-mediated CNS disease, atypical EAE does have advantages, as disease can be induced, is self-limiting, stable, and does not progress to humane endpoints like JME. However, there are distinct differences, specifically atypical EAE is associated with robust IgG response to conformational MOG epitopes, whereas JME exhibits positive CSF finding that have not yet been further evaluated for myelin specificity.3, 12 Interestingly, although we isolated and detected JMRV in JME lesions, we have not detected anti-JMRV antibodies in the CSF, implying that CSF IgG targets other antigens. Importantly, the most distinct difference we report here is JME possesses T-cell responses to myelin-specific antigens that were not induced by immunization, demonstrating that JME possesses features that make it a spontaneous IDD model for MS.
In conclusion, animal models that share clinical, immunological, and pathological aspects of human disease are valuable for assisting in determining etiological agents, mechanisms, and treatment options. We have found that some animals with JME possess myelin-reactive T cells in their lesions and these were predominately comprised of CD4+ T cells with Th1 and Th17 phenotypes, and fewer CD8+ T cells with CTL and Tc17 phenotypes, which are both considered to play a proinflammatory pathogenic role in MS. These novel findings draw further parallels between JME and MS, and demonstrate that JME can serve as an outstanding NHP model to investigate mechanisms that lead to an IDD. Our data also indicate that a potential trigger of JME, JMRV, does not itself induce myelin-reactive T cells, supporting the hypothesis that JME and possibly MS are triggered by mechanisms involving myelin damage and not myelin epitope mimicry.35 Future studies will focus on the investigation of triggers that precipitate JME development and progression, as understanding how disease initiates could lead to discovery of mechanisms driving MS.
Acknowledgments
The authors thank the dedicated animal care staff at the ONPRC for the humane treatment of the animals exhibiting JME, and Drs. Ben Burwitz, Jonah Sacha, and Andrew Sylwester for assistance with IFNγ ELISpot analysis, and ICS and FCM, respectively. This research was supported by the United States Department of Defense grants [W81XWH-09-1-0276 (LSS, BF, SGK, WDR, and SWW) and W81XWH-17-1-0101 (LSS and SWW)], National Institutes of Health grants that support the ONPRC Flow Cytometry and Japanese Macaque Resource [P51OD011092 (MKA, BF, SGK, LSS, and SWW)] and R24-NS104161 (LSS, BF, SGK, and SWW), The Laura Fund for Multiple Sclerosis Research (DNB and SWW), and the Race to Erase MS Center Without Walls (SWW).
Conflict of Interest
All authors have nothing to report.
Author Contributions
ANG and KSF designed and performed the ICS and FCM, and IFNγ ELISpot analyses, processed the data, and drafted portions of the manuscript. MM performed the histopathological analysis and created information for Table 1. IT and WDR developed MRI protocol and analyzed the scans to detect lesions. SGK was responsible for dissection of inflammatory lesions for isolation of MNC infiltrates. GSW was involved in the setup and execution of the JMRV ELISA analyses with ANG. BF and SP were involved in the immunogenetic phenotyping and identifying animals suitable for the study cohorts. MKA, DNB, LSS, and SWW were responsible for the conception and design of the study, and drafting of the manuscript.




