Mitochondrial encephalopathy Due to a Novel Pathogenic Mitochondrial tRNAGln m.4349C>T Variant
Abstract
Objective
Mitochondrial diseases are a group of genetic diseases caused by mutations in mitochondrial DNA and nuclear DNA, among which, mutations in mitochondrial tRNA genes possessing prominent status. In most of the cases, however, the detailed molecular pathogenesis of these tRNA gene mutations remains unclear.
Methods
We performed the clinical emulation, muscle histochemistry, northern blotting analysis of tRNA levels, biochemical measurement of respiratory chain complex activities and mitochondrial respirations in muscle tissue and cybrid cells.
Results
We found a novel m.4349C>T mutation in mitochondrial tRNAGln gene in a patient present with encephalopathy, epilepsy, and deafness. We demonstrated molecular pathomechanisms of this mutation. This mutation firstly disturbed the translation machinery of mitochondrial tRNAGlnand impaired mitochondrial respiratory chain complex activities, followed by remarkable mitochondrial dysfunction and ROS production.
Interpretation.
This study illustrated the pathogenicity of a novel m.4349C>T mutation and provided a better understanding of the phenotype associated with mutations in mitochondrial tRNAGln gene.
Introduction
Mitochondrial diseases are common neurologic disorders caused by mitochondrial DNA (mtDNA) or nuclear DNA (nDNA).1 With the application of next generation sequencing(NGS), more and more mtDNA variants have been reported to be associated with mitochondrial diseases.2 However, due to the very high sequence diversity of mtDNA, evaluation of the pathogenicity of these variants are usually complicated.3 Hence, many methodological approaches have been described to evaluate the pathogenicity of a novel mtDNA mutation.4-7
Hundreds of mtDNA variants associated with a variety of human diseases have been reported in the updated human mitochondrial genome database-MITOMAP,8-10 only 93 variants have a confirmed status, whereas other 676 variants are classified as reported. Of the 93 confirmed pathogenic variants, more than half of them (54%) are located in the mitochondrial tRNA genes and only a few occur at the anticode bases of the tRNA which are critical for code recognition.11 This suggests that this key site mutation may lead to severe phenotypes and incompatibility with embryonic development and postnatal development. In contract, pathogenic mutations causing milder phenotypes are likely compatible with embryonic and postnatal development.12 However, most of the molecular pathomechanisms of these relative benign mutations are highly complex and far from well-documented and hence, the effective therapies remain limited. It is therefore very important to elucidate how these point mutations directly affect the structure and function of mitochondrial tRNAs.
Herein, we reported a 12-year-old body with mitochondrial disease harboring a novel m.4349C>T mutation in mitochondrial tRNAGln gene. We confirmed the pathogenicity of this mutation by a combination of different methods, including clinical evaluation, muscle histochemistry, northern blotting analysis of tRNA levels, biochemical measurement of respiratory chain complex activities, and mitochondrial respirations in cybrid cells with this mutation.
Patients and methods
Clinical evaluation
Figure 1A shows the pedigrees of the family with a novel m.4349C>T mutation. The proband was a 12-year-old male born from a healthy and nonconsanguineous Chinese couple after a normal pregnancy and delivery. His developmental milestone was mildly retarded with walking by himself at 15 months, but not steadily as his peers. He frequently complained of vomiting without any causes since 3-year-old. At the age of six years, he began suffering from sensorineural deafness and recurrent headache. He presented with occasionally seizure attacks since 7 years old and had been taking antiepileptic drugs since then. Family history was not contributory. Physical examination at the age of 12 years old revealed a height of 128cm and a weight of 17 kg, with hypertrichosis in lower limbs. His muscle strength and deep tendon reflexes were normal. The Babinski sign and signs of meningeal irritation were negative. Other physical examination did not reveal any abnormalities. Laboratory examinations showed a normal lactate acid in blood at rest (1.30 mmol/L, normal <1.55 mmol/L). Brain magnetic resonance imaging (MRI) demonstrated obvious brain atrophy (Fig. 1B).
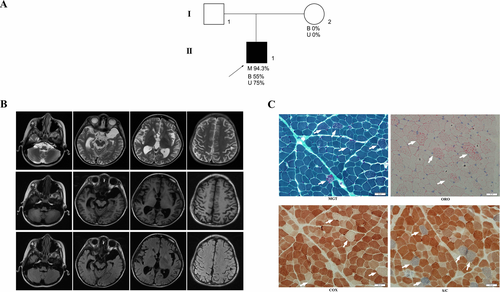
Muscle histopathological studies
Muscle biopsies were performed and histochemical studies were carried out. Serial cryosections, 8µm in thickness, were stained with hematoxylin–eosin(HE), modified Gomori trichrome (MGT), NADH-tetrazolium reductase (NADH), Succinate reductase (SDH), Cytochrome C oxidase (COX), Succinate reductase/Cytochrome C oxidase (S/C), Oil Red O(ORO), and Adenosine triphosphatase (ATPase pH 9.4, 4.6, 4.3) as previously described.13
Genomic analysis
Total DNA was extracted from blood, urinary sediment, and muscle biopsy specimens using a standard procedure as previously described.14 The entire mtDNA sequences of proband and his mother were obtained from the NGS. The quantitation of the heteroplasmy of m.4349C>T mutation was confirmed by Pyosequencing as described previously.15, 16
Cell line and culture condition
The 143B TK– cell lines were grown at 37°C in a humidified 5%CO2 atmosphere in DMEM, containing 4.5 g/L glucose, 10% (V/V) fetal calf serum, 1 mmol/L sodium pyruvate, 200 U/mL Penicillin G, 200 mg/mL streptomycin, 110mg pyruvate per L, and 100μg/mL BrdU. The ρo cell lines(human cells lacking mtDNA), deriving from 143B TK–, were grown in DMEM with the addition of 50 μg/mL uridine. The mutant and wild-type (WT) mtDNA derived from patient’s platelets was intercellularly transferred into ρo cell, as detailed previously.4, 17 Cybrid cell lines with more than 95% mutant mtDNA (MUT) and its absence of this mutation(WT) were used for the experiments. All cybrid cells were maintained in the same medium as the 143B TK– cell lines.
Western blots analysis
Muscle biopsies and cybrid cells were lysed in cell lysis buffer (Beyotime, China). 30-50 mg protein were run on a 8-12% sodium dodecyl sulfate-polyacrylamide gel (SDS–PAGE) at 100 V, 2-3 h. Proteins were transferred to PVDF (Immobilon-P PVDF-Membrane, Millipore, Burlington, MA, USA) for 1-3 h at 100 V, 4°C. After blocking with 5% dried skimmed milk, the membrane was incubated with various primary antibodies. Primary antibodies were as follows: total OXPHOS antibody cocktail (ab110413, Abcam), anti-CO4 (CST 4805s, Cell Signaling Technology), anti-GAPDH (10494-1-AP, Proteintech), anti-ND5 antibody(ab92624, Abcam), anti-ATP6 antibody (ab192423, Abcam), anti-ND1 antibody (ab181848, Abcam), Anti-CYB antibody (ab81215, Abcam), Anti-VDAC1/Porin antibody (ab158995, Abcam), Anti-β actin antibody (TA-09, Zhongshan Jinqiao).
Measurement of oxygen consumption rate
The oxygen consumption rate (OCR) measurements in the cybrid cells were performed using a Seahorse Bioscience XFe24 instrument ( Agilent, Santa Clara, CA, USA), as detailed previously[22]. Cybrid cells were seeded at a density of 4 × 104 cells per well on Seahorse XFe24 polystyrene cell culture plates. The OCR in the permeabilized cybrid cells were also analyzed by a Seahorse Bioscience XFe24 instrument, as detailed described previously.18
Spectrophotometric assays of respiratory chain enzyme activity
Mitochondria from cybrid cells were isolated as previously described.19, 20 The isolated mitochondria were immediately stored at −80˚C until carrying out the biochemical analysis. Spectrophotometric determinations of enzyme activity include NADH-ubiquionne oxidoreductase (complex I), succinate dehydrogenase (complex II), cytochrome c oxidase (complex IV), and citrate synthase.
Assessment of mitochondrial ROS production
The levels of mitochondrial reactive oxygen species (ROS) production were determined using MitoSOX assay as detailed previously.21, 22 Briefly, approximately 1 × 106 cybrid cells were cultured with 1 µmol/L MitoSox for 20 min at 37°C, washed twice and resuspended in PBS, and analyzed by flow cytometry (excitation at 510 nm and fluorescence detection at 580 nm).
Mitochondrial membrane potential (ΔΨm)
The ΔΨm in cybrid cells was performed by using JC-10, a mitochondrion selective dye, which changes its color from green to red reversibly as membrane potentials increase. Cybrid cells were grown on glass coverslips for 24 h, incubated with 10 µg/mL JC-10 for 20 min and examined by confocal microscope (Leica LCS SP8).
Statistical analysis
Quantitative data are represented as the mean ± SEM. The independent samples t-test was used to compare mutant and WT cybrid cells. The statistical software GraphPad Prism and SPSS 23.0 statistical software were used for data analysis. P < 0.05 was considered statistically significant.
Results
Histopathological study
Muscle pathologies of the patient showed a great prevalence of ragged-red fibers (RRFs), abundant fibers with lipid storage, and numerous COX-negative fibers, which are strongly suggestive of mitochondrial diseases (Fig. 1C).
Molecular genetic analysis
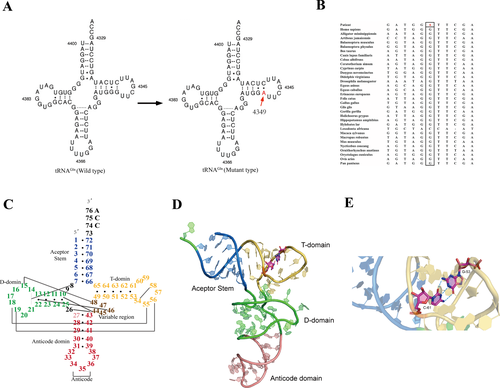
The complete mitochondrial genome analysis revealed a heteroplasmic m.4349C>T mutation in the mitochondrial tRNAGln gene. The heteroplasmic rate of the m.4349C>T mutation in the blood(B), urinary sediment(U), and muscle samples were 55%, 75%, and 95.5%, respectively. However, we failed to detected this mutation in the blood and urinary sediment samples from his mother. This mutation resulted in inappropriate base pair interaction, from tightly G-C to A-C mismatching, in the T-stem of tRNAGln (Fig. 2A). Multialignment analysis of the tRNAGln showed highly evolutionary conservation of the nucleotide at this position (Fig. 2B). According to the second structure of which the position of nucleotides were numbered by Sprinzl et al,23 the novel m.4349C>T mutation abolished the last base-pairing (53–61) of T-stem of the classical tRNA (Fig. 2C). The A-C mismatching results in an opening of the T-stem and is predicted to be favorable for instability of T-loops (Fig. 2D and E).
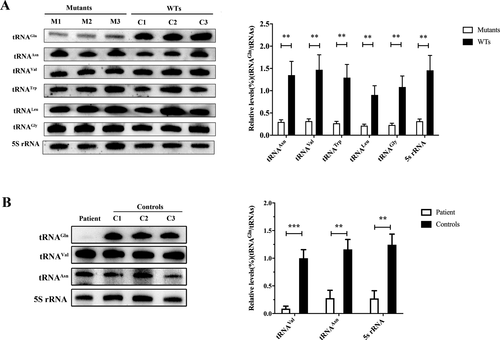
The m.4349C>T mutation contributes to a reductions in the steady-state level of tRNAGln
To examine whether the m.4349C>T mutation affects the steady-state level of tRNAGln, total RNA from cybrid cells, as well as muscle samples were subjected to Northern blots hybridizing with DIG-labeled oligo-deoxynucleotide probes for tRNAGln, tRNAAsn (for the light strand transcription), tRNATrp, RNAGly, tRNAVal, and tRNALeu(UUR)( for the heavy-strand transcription unit),21, 24 and 5s rRNA (for nDNA transcription), respectively. As shown in Figure 3A, the amounts of tRNAGln in cybrid cells were remarkable decreased, compared with those of WT cybrid cells. The average level of tRNAGln in the mutant cybrid cells were decreased by 77.94%, 78.5%, 79.32%, 76.32%, 78.64%, and 78.5% of the average values of WT cybrid cells after normalization to tRNAAsn, tRNAVal, tRNATrp, tRNALeu(UUR), tRNAGly, and 5s rRNA, respectively (Fig. 3A). Furthermore, the average level of tRNAGln in patient’s muscle sample was also dramatically decreased by 91.12%, 76.03%, and 78.08% compared with controls after normalization to tRNAVal, tRNAAsn, and 5s rRNA (Fig. 3B).
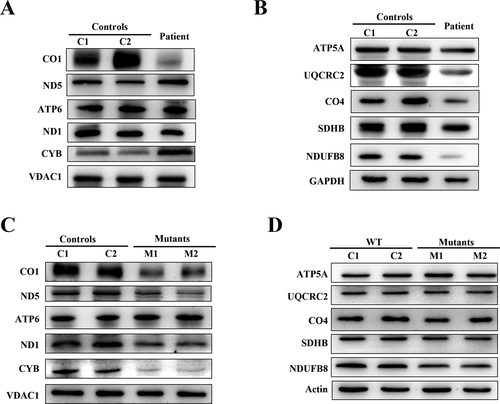
The m.4349C>T mutation leads to decreases in mitochondrial proteins
We then examined the levels mtDNA-encoded subunits of oxidative phosphorylation (OXPHOS) complexes in patient’s muscle tissue and cybrid cells by Western blots analysis using antibodies binding to CO1, ND5, ND1, ATP6, and CYB. Furthermore, some of nDNA-encoded subunits of OXPHOS complexes in muscle tissue and cybrid cells were also detected by Western blots analysis using antibodies against five polypeptides (NDUFB8 of complex I, SDHB of complex II, UQCRC2 of complex III, CO4 of complex IV, and ATP5A of complex V). As shown in Figure 4A, the expression levels of CO1 and ND1 was dramatically decreased in the muscle sample. On the contrary, the CYB expression was also significantly increased. The nDNA encoded UQCRC2, CO4, and NDUFB8 were all significantly decreased (Fig. 4B). Different from muscle specimens, the expression levels of CO1, ND1, and CYB in mutant cybrid cells were all remarkable decreased (Fig. 4C). However, of the nDNA-encoded proteins, only NDUFB8 expression was mildly decreased in mutant cybrid cells (Fig. 4D).
The m.4349C>T mutation results in mitochondrial respiration deficiency
To further access whether the m.4349C>T mutation affects cellular bioenergetics, we determined the bioenergetics profile of mutant and WT cybrid cells using a Seahorse Bioscience XFe24 Extracellular Flux Analyzer.
The basal oxygen consumption rate (OCR) was determined by the first three rates. Then oligomycin, FCCP, antimycin A, and rotenone were added in turn to measure the respiration of mitochondria and nonmitochondria. The difference between the basal OCR and the oligomycin-sensitive OCR produces the amount of ATP-linked respiration. The basal respiration in the mutant cybrid cells was 72.4% relative to the WT cybrid cells. The ATP-linked respiration and maximal respiration in mutant cybrid cells were 70.5% and 60.8% relative to the mean value measured in WT cybrid cells, respectively (Fig. 5A). These results suggested that m.4349C>T mutation significantly decreased energy expenditure and mitochondrial oxidative capacity.
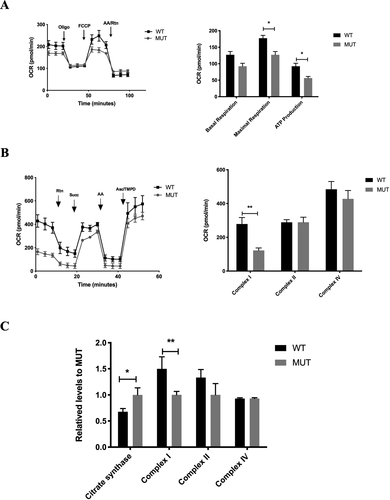
To further identify site-specific changes of the respiratory chain, we measured complex I-, II-, and IV-mediated respiration in permeabilized cybrid cells. With the initial presence of malate and pyruvate (the substrate for complex I), then sequential additions of rotenone (to inhibit complex I), succinate (the substrate for complex II), antimycin A (to inhibit complex III), and ascorbate plus TMPD (the substrate for complex IV) are injected to determine mitochondrial respiration. As shown in Figure 5B, although, m.4349C>T mutation did not affect the complex II- and IV- mediated respiration, however, the complex I-mediated respiration was remarkably decreased by 56.1% relative to the WT cybrid cells.
We further use biochemical assay to determine the activities of complexes of mitochondrial respiratory chain. Consistently, the mutant cybrid cells showed a significant 33.3% reduction in the activities of complex I compared with WT cybrid cells. However, the citrate synthase activity in mutant cybrid cells was mildly elevated by 32.2% relative to WT cybrid cells (Fig. 5C).
The m.4349C>T mutation results in elevated mitochondrial ROS production and collapse of mitochondrial membrane potential
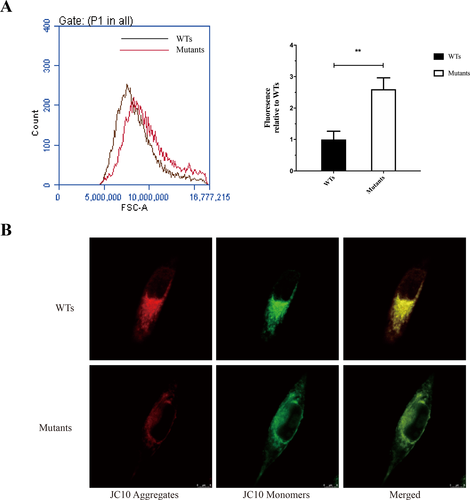
The levels of mitochondrial ROS generation in mutant and WT cybrid cells were determined using MitoSOX assay via flow cytometry. As shown in Figure 6A, the levels of ROS generation in the mutant cybrid cells showed significantly 2.6-fold higher than that of WT cybrid cells. We further measured the ΔΨm in cybrid cells using JC-10, a mitochondrion selective dye, which changes its color from green to red reversibly as membrane potentials increase. Staining and imaging analysis showed that JC-10 formed red fluorescent aggregates in WT cybrid cells, whereas JC-10 existed in monomeric form and stained cells green (Fig. 6B).
Discussion
In this study, we identified a novel pathogenic m.4349C>T mutation in a 12-year-old boy with encephalopathy, epilepsy, and deafness. A number of features suggested this novel mutation to be pathogenic. First, multialignment analysis showed highly evolutionary conservation of this nucleotide. Second, the m.4349C>T mutation was heteroplasmic among the available tissues, with a higher mutation load in muscle and a less extent in blood cells. Third, muscle pathologies showed a histochemical evidence of a mitochondrial disorder, such as COX-deficiency fibers, RRFs, and lipid storage fibers. Fourth, mutant tRNAGln steady-state level was both dramatically decreased in muscle and cybrid cells. Fifth, remarkable mitochondrial respiratory deficiency was noted in the mutant cybrid cells. According the pathogenicity scoring system,5, 25 the m.4349C>T mutation is definitely pathogenic. The deleterious effects of tRNAGln m.4349C>T mutation contributed to pathogenesis can be conceivably hypothesized that the inappropriate base pair interaction alters tRNAGln structure and disturbs the synthesis of mtDNA-encoded respiratory chain complexes subunits and in turn, markedly results in widespread mitochondrial dysfunction.
To date, there are 10 reported disease-linked variants have been described in mitochondrial tRNAGln gene (MITOMAP: A human mitochondrial genome database, http://www.mitomap.org), of which the m.4332G>A mutation was the only confirmed pathogenic mutation in tRNAGln gene according to the criteria outlined in previous studies.5, 25 There are only two patients reported to harbor this mutation, both of whom presented with encephalopathy and deafness.26, 27 The patient in this present study also presented with encephalopathy, epilepsy, and sensorineural deafness since 6 years old. The m.4369A>AA and m.4372C>T were another possible pathogenic mutations in tRNAGln gene according to the MitoTIP tRNA Scoring,28 and both of them located in the anticodon stem. Patient with m.4369A>AA mutation presented with myopathy and progressive external ophthalmoplegia,29 whereas the phenotype of m.4372C>T was not available.30 It seems that encephalopathy and deafness are more common in patients with mitochondrial tRNAGln gene mutations.
The novel m.4349C>T mutation abolished the highly conserved last base-pairing (G53–C61) of T stem of tRNAGln. In a large sequence comparison of the 22 mammal mitochondrial tRNA, the 51–63 base pair of tRNAGln was 100% GC base pair.31 Of the 46 E. coli tRNAs, 26 have the 51–63 base pair, which is expected to give tighter binding32 and confer the specificity for binding by Elongation Factor-Tu(EF-Tu).33 Hence, it could conceivably be hypothesized that the m.4349C>T mutation loses the appropriate base pair interaction, from tightly G-C to A-C mismatching, in the T stem of tRNAGln and, in turn, alters the structure and function of tRNAGln. Consistently, Northern blots analysis showed marked reduction in the levels of steady-state tRNAGln in patients’ muscle and mutant cybrid cells. These data indicate that m.4349 C>T mutation decreases the tightest binding of 51-63 base pair and make it to be easily to be degraded.
Mitochondrial tRNAs are important for the accurate translation the subunits of respiratory chain complexes (I, III, IV, and V). Therefore, mutations typically lead to combined respiratory complex deficiencies.34 In the present case, mutant mitochondrial tRNAGln predominantly disturbs the complex I and IV subunits as demonstrated by significant decrease in CO1, ND1, CYB, and NDUFB8 protein levels, and lead to combined respiratory complex deficiencies, especially complex I and IV deficiency as demonstrated by remarkable COX deficiency fibers in muscle histopathology and significant complex I-mediated respiratory and activity deficiency in the mutant cybrid cells. However, we did not note a significant decrease in the activity of complex IV and its mediated respiration in the mutant cybrid cells. In fact, all the confirmed pathogenic mutations in tRNAGln gene are reported to have evidence of COX deficiency26, 27 in muscle histopathology. It is interesting that respiratory chain complexes activities and subunits expression levels were different between muscle sample and cybrid cells, which may be due to cell or tissue type-specific physiological impairment.35, 36 Therefore, our findings clearly demonstrate m.4349C>T mutation generally lead to combined respiratory complex deficiencies.
The impairment of mtDNA translation caused by m.4349C>T mutation led to dysfunction of mitochondria. The ATP-linked and maximal respiration were significantly decreased in the mutant cybrid cells compared with those of WT cells. As we know, the maximal respiration may be used by cells to protect mitochondrial function from oxidative insults.37, 38 As mentioned in the literature review,39, 40 the impairment of mitochondrial respiratory lead to more electron leakage and in turn, elevate the oxidative stress level. Consistently, ROS production in cybrid cells carrying this mutation was significantly increased, whereas the mitochondrial membrane potential was remarkably decreased compared with those in WT cybrid cells. Increased oxidative stress level results in collapse of the mitochondrial membrane potential, and finally, premature cell senescence when a certain threshold level is reached.41, 42
Thus, we reported a novel pathogenic m.4349C>T mutation in mitochondrial tRNAGln gene and demonstrated molecular pathomechanisms of this mutation. The m.4349C>T mutation resulted in impairment of translation machinery of mitochondrial tRNAGlnand in turn, causes remarkable mitochondrial dysfunction.
Acknowledgments
The authors thank the patient and his families. This study was supported by the National Natural Science Foundation of China (No.81701237 and 81671235), People's benefit project of science and technology in Qingdao (16-6-2-1-nsh), and the Taishan Scholars Program of Shandong Province.
Authors' contributions
K.J and C.Y designed the studies. C.Y and Y.Z supervised the work. W.W, X.X, and Y.L performed the experiments. K.J and D.W analyzed the data and wrote the manuscript. F.L and C.Y reviewed the manuscript and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors have declared that no conflict of interest exists.




