Sugarcane
Abstract
As more than 70% of the sugar harvested for human consumption is derived from sugarcane, improving sucrose content, biomass yield, and resistance to pests and diseases remains an important focus of traditional breeding programmes. In addition, genetic engineering research has supported the introduction of specific traits and facilitated further understanding of complex physiological pathways in the plant. Transgenesis has allowed diversification of output traits so that a range of sugars, biopolymers, neutraceuticals, industrial enzymes, and pharmaceuticals can be produced by the plant. Sugarcane is considered as a critical component of our bioenergy future and is currently a major feedstock for ethanol production. Significant advances in associating putative biological functions to sugarcane genes have been achieved by the Brazilian SUCEST project. Future genetic improvement of sugarcane will rely on a better understanding of metabolic control and flux, cellular compartmentation and availability of metabolites, and the ability to identify potential and crucial targets for genetic engineering.
1 Introduction
1.1 History, Origin, and Distribution
Sugarcane is a large grass of the genus Saccharum, tribe Andropogoneae, family Poaceae. Modern sugarcane (Saccharum spp.) cultivars are interspecific hybrids derived from a hybridization process involving Saccharum officinarum (or “noble cane”) and Saccharum spontaneum (wild cane), followed by a series of backcrosses to the noble parent (Daniels and Roach, 1987).
The earliest known historical record of sugarcane and sugar is from Indian writings from 3000 to 3400 years ago. The generic name for sugarcane, Saccharum, originated from the Indian Sanskrit term “sharkara” for the crude sugary product obtained from the honey reeds. Dispersal of Indian sugarcane westward seems to have occurred during the first millennium BC. Soldiers of Alexander the Great are known to have carried it to Europe from India about 325 BC. Later, Greek and Roman writers were familiar with the concept of the Indian honey reed and its “honey” (sugar) product. The early history of sugarcane is covered by a number of authors, including Deer 1949 and Barnes 1964.
The origin of sugarcane is a complex question that is best discussed in relation to its taxonomy and distribution in Southeast Asia, the Indonesian Archipelago, and New Guinea. Different species likely originated in various locations with S. officinarum and Saccharum robustum in New Guinea, Saccharum barberi in India, and Saccharum sinense in China. Dispersal of S. officinarum over a period of thousands of years is believed to have occurred both into the Pacific Ocean area, and along the island chain into Asia, whilst the thinner Indian canes were developed and cultivated in the North India/South China region.
Initially, pieces of cane stalk would have been chewed to express the sweet juice, and chewing canes still provide a conveniently packaged form of energy food in many cultures. Juice extraction from the stalk, and concentrating it by drying or heating to produce a crude sugary product, must have been developed in a rudimentary form at least 3000 years ago. The art of sugar manufacture took longer to develop, probably in India and perhaps less than 2000 years ago. Deer 1949 considered that Nestorian Christian monks at the mouth of the Euphrates river were the first to refine the crude raw product into a form of “white” sugar about 450 AD.
The Mediterranean sugar industry was the first major one in Europe, and began about the time of the Arabian conquest of Egypt in 640 AD. They spread it across North Africa and into Spain by 750 AD, where it was important for many years, with 30000ha under cane by 1150 AD. By the early 1500s AD, cane was carried by the Spanish to the Caribbean and the Americas, and by the Portuguese to West Africa and Brazil. And so the worldwide sugarcane industry was born.
Prior to the 20th century, the world sugarcane industry was dependent on the noble canes (S. officinarum) and the cane from India and China (S. barberi and S. sinense, respectively) (Sreenivasan et al., 1987). These canes were characterized by high sucrose levels and low fiber contents, but were susceptible to several pests and diseases, notably sereh disease (Arceneaux, 1965). Sugarcane breeding and selection became a directed, ongoing process following the observation in 1858 that sugarcane panicles produce viable seed (Stevenson, 1965). The Dutch established a breeding and selection program in 1888 in Java to incorporate the disease resistance, hardiness, and tillering capacity of S. spontaneum into S. officinarum germplasm. Interspecific crosses were made between S. officinarum and the wild S. spontaneum (which was resistant to sereh) (Stevenson, 1965). The resultant hybrids were continually backcrossed to S. officinarum in a process called nobilization (Stevenson, 1965). This effort resulted in the release of the first of the nobilized hybrid cane cultivars, POJ2725 and POJ2878 in 1921. These two early cultivars served as the foundation in the pedigree of nearly all locally developed and adapted modern sugarcane cultivars worldwide (Moore, 2005). Nobilization became established as a method of retaining the desirable qualities of S. officinarum, retaining the hardiness and disease resistance of S. spontaneum, while diluting the negative effects of wild germplasm (Berding and Roach, 1987).
Currently, sugarcane is widely grown for sugar production in many tropical and subtropical countries (with a minimum of 600mm of annual rainfall) in South, Central, and North America, the Caribbean, Africa and adjacent islands, Southern Asia and Australasia.
1.2 Evolution, Taxonomy, Cytological Features, and Genome Size
Mukherjee 1957 coined the term “Saccharum complex” to describe a large, closely related genera (Erianthus sect Rhipidium, Miscanthus, Sclerostachya, Narenga), which are considered to be involved in the evolution of cultivated species of Saccharum. The phylogenetic relationships within the Saccharum complex have been debated for many years (Daniels and Roach, 1987; Irvine, 1999). A long discourse on how the various species may have evolved is provided by Daniels and Roach 1987.
-
S. officinarum L.: sweet, juicy, thick stalk garden cane, initially in New Guinea
-
S. barberi Jesw.: sweet, thin stalk Indian canes
-
S. sinense Roxb.: sweet, thin stalk Chinese canes
-
S. edule Hassk.: edible inflorescence garden cane, New Guinea, Melanesia
-
S. spontaneum L.: very thin, hardy wild canes, low sugar, New Guinea and southern Asia
-
S. robustum Brandes & Jeswiet ex Grassl: tall, harder, thick stalk wild canes, a little juice and sugar, New Guinea and eastern Indonesia
Modern commercial sugarcane cultivars are highly heterozygous, complex polyploid, and aneuploid hybrids, often with four of the above species of Saccharum in their ancestry. Cytological studies showed that nobilization is characterized by asymmetric chromosome transmission (Bremer, 1961). In a cross between S. officinarum (2n = 80) as the female parent and S. spontaneum (2n = 40–128) as the male parent, S. officinarum generally transmits two haploid chromosome sets while S. spontaneum transmits one. This 2n+n transmission is continued up to the second backcross. A consequence of this is that modern cultivars have chromosome numbers ranging from 2n = 99–130 (Bremer, 1961). In addition, commercial sugarcane cultivars have complex polyploid (10–12 copies of the genome) and aneuploid (100–120 chromosomes) genomes. In polyploids such as sugarcane, the haploid chromosome number (1C value = n) is not the same as the monoploid number (= x) (Butterfield et al., 2001). The monoploid genome size for S. officinarum (x = 10) is approximately 926Mbp (mega base pair), and for S. spontaneum (x = 8) 760Mbp (Butterfield et al., 2001). This base genome number is roughly double the monoploid genome size of rice (415Mbp) and similar to that of Sorghum bicolor Moench (760Mbp) (Butterfield et al., 2001).
1.3 Economic Importance: Production, Utilities, Economic Attributes, and Industrial Uses
Sugarcane is cultivated for its high rate of sucrose accumulation, ease of propagation via vegetative stem cuttings and multiple harvests from a single planting. It is a principal crop in tropical and subtropical regions, with a production estimate of over 1.3 million metric tons of sucrose per annum. It provides approximately 70% of the world's sugar (FAO, 2006).
Sugarcane has long been recognized as one of the most efficient crops in terms of converting solar energy into biomass (Alexander, 1973). It is one of the most effective photosynthesizers in the plant kingdom, able to convert 2% of incident solar radiation into plant biomass. It is the second largest contributor (10–12%) of dietary carbohydrate to humans after the cereals. Sugar processing meets the needs of both high-income consumers (e.g., refined white and specialty sugars) and low-income domestic consumers (e.g., the production of jaggery in India or panela in Colombia, where cane juice is boiled to make cakes of brown sugar). By-products of sugar milling such as bagasse, molasses, furfural, furfuryl alcohol, dextran, and diacetyl (O'Reilly, 1998) have several uses. For example, bagasse (a fibrous residue after sugar extraction) can be used to fuel boilers in the sugar mills, to generate electricity for the local power grid, to manufacture paper, and as an animal feed. Molasses are used in syrups and animal feed and as a substrate for ethanol production.
Sugarcane is considered as a critical component of our bioenergy future as: (i) sugarcane is already used in the production of ethanol, produced by fermentation and distillation of sugars. Currently, Brazil is the world's largest producer of sugarcane ethanol (Moreira, 2000). Ethanol can be blended with gasoline (gasohol) or diesel (biodiesel or dieselhol); (ii) production of energy, such as ethanol, from sugars is more efficient than production from grains, in both cost for joule produced and energy input/output efficiency. For each unit of fossil energy input to the sugarcane agro-industrial system, nine units of renewable energy output (ethanol plus surplus bagasse) result, compared to less than 2 units resulting from grains, such as maize; and (iii) sugarcane is ranked first among all other crops for biomass production (FAO: http://www.fao.org; Moreira, 2006). As a perennial crop, it has more advantages for biofuels production than annual crops. It is more efficient at solar energy conversion, and it can be harvested annually for a number of years without replanting.
1.4 Traditional Breeding: Breeding Objectives, Tools and Strategies, and Achievements
The objectives of sugarcane breeding programs around the world are to produce cultivars with improved characteristics such as increased cane yield, higher sucrose content, pest and disease resistance, tolerance to abiotic stress, and improved ratooning ability. Focused breeding programs have led to significant contributions to characters such as those listed above (Hogarth, 1976; Nuss, 2001).
Crossing of two parents is the first step in producing and selecting a new cultivar. If pollen and seed production do not occur naturally, as is the case in sugarcane growing areas with latitudes above 15° north and south, crossing is conducted in photoperiod glasshouse facilities where day length and temperature can be manipulated (Brett, 1974). Extensive evaluation of the progeny from the crosses is undertaken. The selection process consists of several different stages and usually takes 10–14 years to release a variety. This prolonged period before a new sugarcane variety can be commercially released is largely due to the reliance of the selection process on phenotypic characters. At each stage, clones with unsuitable characters are discarded and the performances of selected clones are evaluated in larger plots (Parfitt, 2005).
Due to the narrow genetic base of modern varieties (Hogarth, 1987), sugarcane breeders have tried to exploit the genetic variation within the Saccharum complex, which shows great variation for a range of traits of interest to the breeders, such as sugar content, tolerance to drought and cold, pest and disease resistance, uprightness, fiber content, tiller number, stalk size and strength, low suckering, easiness to detrash (suitability for mechanical, green harvest), and ratooning ability. Breeders have used S. spontaneum, Miscanthus sinensis, Erianthus arundinaceus, and Erianthus rockii in introgression initiatives.
S. spontaneum. In the development of modern sugarcane cultivars, an average of only 15–25% of chromatin is derived from S. spontaneum (D'Hont et al., 1995), due primarily to the limited number of S. spontaneum genotypes used (Berding and Roach, 1987). Only two genotypes of S. spontaneum were used in the initial crosses made in the late 19th and early 20th centuries in India and Java (Martin, 1996). S. spontaneum has been considered as a source of positive alleles for traits involved in adaptation to different climatic conditions and for disease and insect resistance. Dunckelman and Breaux 1972 studied the agronomic habits of 32 apparently mosaic-resistant S. spontaneum genotypes to ascertain their potential utilization as breeding material, and found a genotype (US 56-15-8) particularly sweet, with a juice Brix reading of 11.5%. The characterization of sugar composition of S. spontaneum genotypes from the World Collection of Saccharum (Miami, Florida) also indicated that this species is a potential source of positive alleles for sugar content (Tai and Miller, 2001). These results were confirmed by da Silva et al. 2007 in a study involving molecular markers to test if alleles with positive effects for sucrose content could be found in S. spontaneum. Expressed sequence tags (ESTs) involved in sucrose accumulation from the metabolism of complex carbohydrate pathway (da Silva et al., 2007) were used to develop molecular markers. By targeting four functionally characterized sugar metabolism candidate genes to a set of 50 S. spontaneum genotypes showing variation in sugar content, S. spontaneum-specific polymorphic markers were identified. These markers are not present in commercial sugarcane genotypes and may therefore be used for tagging positive S. spontaneum alleles for introgression into commercial sugarcane genotypes. Efforts to introgress these alleles were made in 2005 at the US Department of Agriculture (USDA) Sugarcane Unit in Houma, Louisiana, with a cross involving the commercial cultivar HoCP00-950 and the S. spontaneum genotype MPTH97-216, from Thailand.
M. sinensis. A difficulty in breeding sugarcane for stress tolerance is the trade-off between stress tolerance and yield (Ming et al., 2006). Developing varieties adapted to a wider range of climatic regimes could improve sugarcane production in water-restricted and/or colder regions. Sugarcane with increased water use efficiency and tolerance to drought or cold temperatures are critical selection criteria for that goal. Another way to overcome this difficulty would be to identify alternative alleles contributing to stress tolerance in the Saccharum complex and introgress these into commercial germplasm. Even though the water use efficiency of sugarcane is high, substantial amounts of water are required to maintain maximal growth and productivity. Since irrigation of sugarcane fields is limited, and irrigation of biomass crops is unlikely to be economic, it is important to identify genotypes that are tolerant to water stress. Screening with Paraquat (methyl viologen) for drought tolerance in sugarcane (Ming et al., 2001b), wheat, and barley (Altinkut et al., 2001) has proved to be a rapid and practical screening method, in conjunction with chlorophyll fluorescence measurements, for identifying and characterizing genetic variation in sugarcane water stress tolerance (Ming et al., 2006).
Miscanthus species are exceptionally tolerant to low temperature and drought and are among the few plants in temperate climates that use the C4 photosynthetic pathway (Naidu et al., 2003). In a study comparing drought tolerance of different Miscanthus species (M. sacchariflorus, M. giganteus, and M. sinensis), M. sinensis was the only one that did not show senescence caused by water deficit (Clifton-Brown et al., 2002). M. sinensis retained all of its green expanded leaf area irrespective of water supply, showing its complete resistance to senescence. Similarly, when commercial varieties, breeding lines and seedlings of sugarcane and Miscanthus were exposed to freezing temperatures for at least 2h in the Rio Grande Valley of Texas in 2004, leaf damage was observed on all sugarcane plants, but not on Miscanthus × Saccharum sp. hybrid seedlings (Figure 1). Miscanthus species also have high cellulose fiber content and are considered a potential energy crop by the European Union (EU) (Clifton-Brown et al., 2004).

Miscanthus sinensis × Saccharum sp. hybrid seedlings showing cold tolerance under subfreezing field conditions
If the superior drought and cold tolerance and high fiber of Miscanthus could be combined with the photosynthetic capacity of commercial sugarcane, it would be possible to produce a low-input, high-biomass, drought- and cold-tolerant energy crop. As M. sinensis (2n = 2x = 38) is closely related to sugarcane, it produces viable hybrids when crossed with sugarcane (Grassl, 1967; Lo et al., 1986), and can be seen as a source of stress tolerance genes for introgression purposes. Atienza et al. 2003 have shown that it is feasible to develop a marker-assisted selection program for biomass production in Miscanthus using quantitative trait loci (QTL) to detect markers for traits such as diameter, height, and panicle size. These QTLs could assist in introgression work.
Erianthus species. The Erianthus genus contains eight species (Aitken et al., 2007) and is also a potential donor for stress-resistant genes. E. arundinaceus has several traits desirable to sugarcane breeders such as tolerance to drought and waterlogging, high resistance to Pachymetra root rot, vigor, and good ratooning performance (Berding and Roach, 1987). Many attempts have been made to generate sugarcane × E. arundinaceus hybrids. In situ hybridization analysis of progeny from some of these crosses indicated that the introgressions were successful (D'Hont et al., 1995; Figure 2). However, only one appears to have been successful in producing fertile offspring. Cai et al. 2005 describe the intergeneric hybridization of two populations using the S. officinarum Badilla as the female parent in both crosses with pollen from three different E. arundinaceus clones from Hainan, China. They also describe the backcrossing of the F1 hybrid, YC96-40 with sugarcane cultivar CP84-1198 to generate the backcross 1 (BC1) population, demonstrating the fertility of the intergeneric hybrid. Molecular markers were used to confirm both the introgression into the F1 and BC1 populations and the parentage of the BC1s.
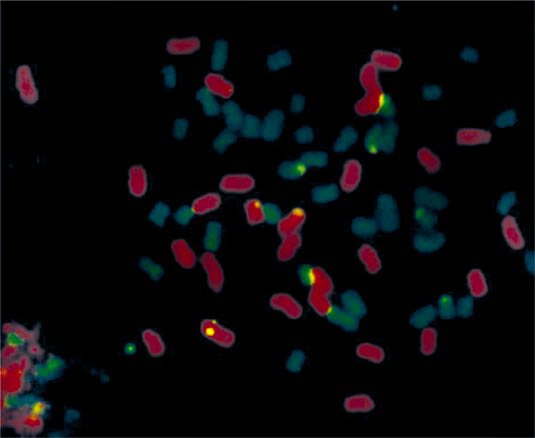
Chromosomes from a root-tip cell of an intergeneric hybrid fluorescing after genomic in situ hybridization. Saccharum officinarum chromosomes are green and Erianthus arundinaceus chromosomes are red [Reproduced with permission from George Piperidis, BSES Limited]
E. rockii. A species originating in the Yunnan, Sichuan, and the Tibetan regions of China, has good vigor, cold and drought tolerance, and good ratooning ability. E. rockii was recently reported by Aitken et al. 2007 to have been successfully used as the male parent for intergeneric hybridization with (1) Vietnam-niuzhe (S. officinarum) and (2) interspecific hybrid Fiji (S. officinarum × S. spontaneum). Using amplified fragment length polymorphisms (AFLP), Aitken et al. 2007 showed that all of 10 screened E. rockii × S. officinarum crosses were true intergeneric hybrids, but that 9 of 10 of the E. rockii × Fiji crosses were selves of the E. rockii female parent and only one was a true introgression. Further analysis showed that there was a (n + n) transmission of gametes.
1.5 Limitations of Conventional Breeding and Rationale for Transgenic Approaches
-
Commercial sugarcane cultivars possess different proportions of chromosomes, complex recombinational events, and varying chromosome sets (aneuploidy) (Sreenivasan et al., 1987). This genomic complexity brings difficulties in applying conventional plant breeding for cultivar improvement. In addition, conventional breeding is a multistage, laborious, and time-consuming process requiring 10–14 years to develop a new cultivar. A single fault, such as disease susceptibility in an otherwise elite cultivar, could cause the cultivar to be abandoned. Conventional breeding approaches to correct such faults in an existing cultivar are impractical in sugarcane, due to the genetic complexity of cultivars (Birch and Maretzki, 1993). The capacity to introduce specific genes by transgenic approach, without major genetic reassortment following crossing, could be used to rescue flawed cultivars (Birch and Maretzki, 1993). For example, the successful production of sugarcane plants resistant to leaf scald disease was achieved by transgenic approach (Zhang et al., 1999).
-
Although breeding efforts in sugarcane have been successful in increasing cane production, only limited success has been achieved recently in increasing sugar content. For example, there has been no increase in sugar content over the last 40 years in Australian sugarcane (Bonnett et al., 2004b). In the USA, Legendre 1995 reported that the average sucrose content of new candidate varieties decreased 3.5% on the fifth cycle of recurrent selection, as compared to the previous cycle, indicating that a limit has been reached for this trait. The QTL analysis of interspecific F1 populations also indicated that modern sugarcane cultivars have a limited (biased subset) population of genes controlling sugar content (Ming et al., 2001b). In contrast, metabolic engineering of sugarcane through transgenic approaches could improve sugar content. For example, transgenic sugarcane with doubled sugar content was achieved when attempting to produce isomaltulose in sugarcane (Wu and Birch, 2007).
-
Production of novel products in sugarcane is not possible by conventional breeding. In contrast, metabolic engineering through transgenic approaches could produce new products, such as alternative sugars, biopolymers, pharmaceuticals, and high-value proteins. For example, successful production of sorbitol (Chong et al., 2007), isomaltulose (Wu and Birch, 2007), p-hydroxybenzoic acid (pHBA) and biodegradable polymer (McQualter et al., 2005; Petrasovits et al., 2007) has been achieved in transgenic sugarcane, which cannot be achieved through conventional breeding.
-
Conventional breeding allows transfer of traits and genes only between sexually compatible species. Hence, transfer of traits from noncompatible species is impossible. In contrast, transgenic approaches allow insertion of novel genes from sexually noncompatible plants/organisms, enable expression of native genes at different levels in specific tissues or under novel developmental patterns of expression.
-
The number of traits to be considered when selecting for variety development is determined by the degree of genetic linkage among those traits. If linkages are rare, several traits can be selected simultaneously. In the case of sugarcane the extent of those linkages is still uncertain (Ming et al., 2006). Recent advances in molecular marker-assisted selection and transformation technologies can alleviate the problem. Thus, genetic transformation by modern molecular techniques (see in Section 2) has the potential to enhance a host of traits including sugar, pest and disease resistance, tolerance to drought and cold, vigor, plant architecture and fiber, and to produce alternative products such as biopolymers and isomaltulose in sugarcane.
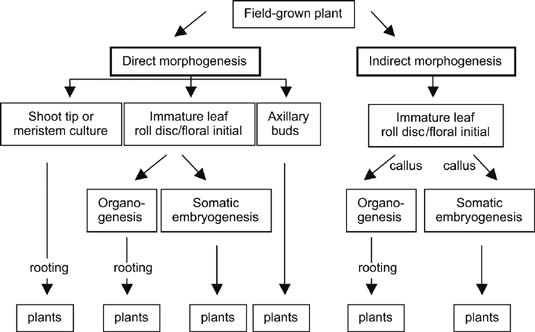 Figure 3
Figure 3Summary of direct and indirect morphogenic pathways for sugarcane regeneration. In the routes where the rooting event is noted, external application of growth hormones is usually required for root production
2 Development of Transgenic Sugarcane
Sugarcane is a prime candidate for the application of genetic engineering, as single characters can be introduced into the complex genetic background of elite commercial clones to correct negative factors, such as disease susceptibility. Successful genetic engineering requires a reliable tissue culture system and efficient transformation methods. Sugarcane was one of the first monocotyledonous crop plants used successfully for establishment of tissue cultures (Barba and Nickell, 1969; Nickell and Maretzki, 1969), regeneration of plants (Heinz and Mee, 1969), and isolation of protoplasts (Maretzki and Nickell, 1973; Nickell and Heinz, 1973).
2.1 Tissue Culture and Transformation
The early established robust tissue culture/regeneration system of sugarcane became a foundation for efficient genetic transformation of this crop. The explant most frequently used for transformation experiments in sugarcane is embryogenic callus. This callus is produced from the culture of immature leaf whorls immediately above the apical meristem. The whorls are sliced into thin (2–3mm) transverse sections and cultured on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with an auxin (usually 2,4-dichlorophenoxyacetic acid) and sucrose, and subcultured every 3–4 weeks. Prolific compact embryogenic callus is produced within 2 months. However, this varies with different clones and needs to be optimized for the genotype being used.
2.1.1 Regeneration of plants
Sugarcane has a well-established history of in vitro regeneration that began in the late 1960s (Heinz and Mee, 1969). Regeneration of sugarcane can occur via several different pathways (Figure 3) (reviewed by Snyman, 2004; Lakshmanan, 2006; Lakshmanan et al., 2005). The pathways of regeneration by somatic embryogenesis or organogenesis have been well characterized and documented (Ho and Vasil, 1983; Guiderdoni and Demarly, 1988; Taylor et al., 1992). For the regeneration of transgenic plants in vitro, the route of morphogenesis is dependent on the explant targeted for DNA delivery. Criteria for explant choice are: (i) a large number of regenerable cells and (ii) maintenance of regenerative capacity during the selection procedure.
Where embryogenic callus is used as the recipient material for foreign DNA, regeneration is via somatic embryogenesis (Bower et al., 1996; Falco et al., 2000) or organogenesis (Gallo-Meagher and Irvine, 1996). Although transgenic plants have been successfully generated via indirect morphogenesis, limitations include the amount of time taken to regenerate a transgenic plant (36 weeks from DNA delivery to glasshouse transfer) (Bower et al., 1996; Snyman et al., 2000) and the incidence of somaclonal variation as evidenced in agronomic variability when plants are evaluated in the field (Grof and Campbell, 2001; Vickers et al., 2005b).
Alternative tissue targets for transgene delivery have been sought, such as exposed apical meristems from axillary buds followed by shoot morphogenesis (Gambley et al., 1993, 1994; Manickavasagam et al., 2004), leaf roll discs (Snyman et al., 2000, 2001) and pre-emergent inflorescences (Snyman et al., 2006) followed by direct somatic embryogenesis. Although these methods have resulted in transformed plants in a reduced time frame compared to protocols employing indirect embryogenesis, determination of phenotypic fidelity in the field has yet to be published.
2.1.2 Transformation methods
Initial work on sugarcane transformation targeted protoplasts as recipient cells and DNA delivery was via polyethylene glycol (PEG) (Chen et al., 1987) or electroporation (Rathus and Birch, 1992). However, neither technique resulted in the production of transgenic plants, as sugarcane regeneration from protoplasts is notoriously difficult. A significant milestone in sugarcane transformation was achieved when particle bombardment of embryogenic callus was reported and a protocol for recovery of transgenic plants was described (Bower and Birch, 1992). Embryogenic callus proved to be a reliable source of tissue for further refinement of microprojectile bombardment protocols and for the production of transgenic plants in several parts of the world (Bower et al., 1996; Gallo-Meagher and Irvine, 1996; Snyman et al., 1996; Joyce et al., 1998; Ingelbrecht et al., 1999; Falco et al., 2000). In addition, Agrobacterium-mediated DNA delivery was also developed using embryogenic callus as recipient cells (Arencibia et al., 1998; Elliott et al., 1998; Enriquez-Obregon et al., 1998; Liu et al., 2003).
Microprojectile bombardment using a gene gun has been achieved using either the particle inflow gun (PIG) (Finer et al., 1992) or a BioRad device (Heiser, 1993). The principle of this transformation method is to coat either tungsten or gold particles with plasmid DNA containing the gene(s) of interest and to bombard these particles into embryogenic sugarcane callus using pressurized helium combined with a partial vacuum chamber. The first report of successful transformation of sugarcane suspension culture cells using the PIG gun was by Birch and Franks 1991 using the GUS (β-glucuronidase) reporter gene and kanamycin selectable marker gene, although no plants were regenerated. This approach was further refined by Bower and Birch 1992 and is now widely adopted for the production of transgenic sugarcane (Bower et al., 1996; Gallo-Meagher and Irvine, 1993, 1996; Franks and Birch, 1992; Snyman et al., 1996; Joyce et al., 1998; Ingelbrecht et al., 1999; Falco et al., 2000). Transformation efficiencies vary depending on genotype, quality of embryogenic callus, length of time spent in vitro, and selection regime employed, but most laboratories have developed a protocol that suits their requirements.
Explant type |
Transformation method |
Gene |
Selective agent (mgl−1) |
Escapes |
References |
|---|---|---|---|---|---|
Callus |
Biolistic |
NptII |
Geneticin (45) |
Nil |
|
Callus |
Biolistic |
Bar |
BASTA (1-3) |
Yes |
Gallo-Meagher and Irvine, 1996 |
Callus |
Agrobacterium |
Hph |
Hygromycin (25) |
Yes |
Arencibia et al., 1998 |
Callus |
Agrobacterium |
Bar |
Bialophos (1) |
Yes |
Elliott et al., 1998 |
Immature leaf whorls |
Agrobacterium |
Bar |
PPT (4) |
Yes |
Enriquez-Obregon et al., 1998 |
Axillary meristem |
Agrobacterium |
Bar |
BASTA (5) |
Yes |
Manickavasagam et al., 2004 |
Callus |
Agrobacterium |
NptII |
Paromomycin (150) |
Nil |
Joyce et al., 2006a |
The desire to minimize transgene copy number and integration complexity, coupled with advances reported in Agrobacterium-mediated transformation in other monocotyledonous crops (reviews by Cheng et al., 2004; Shrawat and Lorz, 2006; Wang and Ge, 2006), made this an appealing system to develop in sugarcane. Although not widely applied yet, Agrobacterium-mediated transformation using embryogenic callus and four different strains of Agrobacterium (Arencibia et al., 1998; Elliott et al., 1998; Enriquez-Obregon et al., 1998; Liu et al., 2003) and axillary buds with two strains (Manickavasagam et al., 2004) has been reported. Although it is difficult to compare transformation efficiencies between the above papers, it appears that pretreatment of the callus prior to Agrobacterium co-cultivation improves transformation efficiency (e.g., a 30min dehydration period). In addition, selection of a particular size of callus (1000μm) (Arencibia et al., 1998), and use of antinecrotic compounds (2mgl–1 silver nitrate; 15mgl–1 ascorbic acid (Enriquez-Obregon et al., 1998) were reported to be advantageous. In both papers, Arencibia et al. 1998 and Enriquez-Obregon et al. 1998, the transgene copy number was 1–3. Elliott et al. 1998 applied no selection pressure for up to 6 weeks after co-cultivation. After this period, GFP (green fluorescent protein)-positive calli were visually selected and transferred to bialaphos at 1mgl–1. Two transgenic plants were regenerated that contained three and seven copies of the GFP gene, respectively.
More recently, transgenic plants were generated using axillary meristems of sugarcane (Manickavasagam et al., 2004). This novel approach resulted in a transformation efficiency of 49.6%. They used two strains of Agrobacterium, LBA4404 and EHA105. The plasmid pGA492 contained the nptII (neomycin phosphotransferase) gene driven by the nos (nopaline synthase) promoter and the bar and gus genes driven by cauliflower mosaic virus (CaMV) 35S promoter. Selection was on 5mgl–1 bialaphos, applied immediately after the co-cultivation period. Transformation efficiency was greatest in both strains when co-cultivation period was for 3 days in the presence of 50μM acetosyringone. Thousands of plants were produced within 5 months. Although chimeric plants were reported, the incidence was eliminated in secondary shoots after five rounds of selection on Basta®-containing medium. When these plants were transferred to the greenhouse and sprayed with herbicide, a total of 336 plants (clones of 10 independent transformation events) representing 50% of total plants screened, displayed herbicide resistance. Southern blot analysis on a small subset of these plants further confirmed the presence of 1–2 copies of the transgene in each plant.
2.1.3 Selection of transformed tissues
Studies on transgenic sugarcane for technology development have involved marker and reporter genes. The reporter genes used include GUS (Jefferson et al., 1987; Gnanasambandam and Birch, 2004; Braithwaite et al., 2004), luciferase (luc) (Mudge et al., 1996b; Gnanasambandam and Birch, 2004), maize anthocyanin regulatory elements (R and C1) (ANT) (Ludwig et al., 1990; Bower et al., 1996; Gnanasambandam and Birch, 2004), and GFP (Elliott et al., 1998; Gnanasambandam and Birch, 2004; Gnanasambandam et al., 2007; Petrasovits et al., 2007). Comparative studies by Bower et al. 1996 on the suitability of GUS, luc, and ANT as reporter genes for transient assays in sugarcane transformation indicated that the ANT system is the most suitable reporter system. ANT expression is visible within 8h after bombardment and steadily increases in intensity up to 48h after bombardment. The expressing cells remained visible in the target tissue for 2–3 weeks, before fading or being overgrown. In addition, the results were not confounded by background ANT activity. In contrast, the main disadvantage of using the GUS reporter gene is that the conditions for detection of gene activity are lethal to plant cells and therefore the transformed event is subsequently lost. Detection of both luc and GFP genes require specialized camera and/or detection systems. In addition, GFP detection is confounded by autofluorescence of the callus and chlorophyll in green plant tissues. However, the power of confocal laser scanning microscopy allows precise visualization of fluorescent signals within a narrow plane of focus, and the reconstruction of three-dimensional structures from serial optical sections (Haseloff et al., 1997). This is an advantage of GFP over both GUS and luc (Gnanasambandam and Birch, 2004).
The most widely adopted stable-integration antibiotic marker used for sugarcane transformation is the aphA2 Escherichia coli Tn5-derived nptII. The first successful application of the nptII-based selection system was reported as a step-wise incremental procedure using geneticin (Bower and Birch, 1992). This formed the foundation for subsequent protocols utilizing geneticin (also known as G418) (Bower et al., 1996; Falco et al., 2000; Snyman, 2004) or paromomycin (Joyce et al., 2006a) as the selective agent (Table 1). Less widely used is the hygromycin phosphotransferase (hph) gene with selection on 20mgl–1 hygromycin (Arencibia et al., 1998; Carmona et al., 2005).
The first paper reporting herbicide resistance in sugarcane also described a selection procedure incorporating a herbicidal agent, bialaphos (Gallo-Meagher and Irvine, 1996). The bar and pat genes from Streptomyces hygroscopicus and Streptomyces viridochromogenes, respectively, encode the phosphinothricin acetyltransferase enzyme that leads to detoxification of phosphinothricin and its derivatives that are ingredients in some commercial herbicides. Selection using herbicides has been used widely, although each laboratory has to determine empirical regimes for selection of different explants and genotypes and formulations of active ingredients differ (Gallo-Meagher and Irvine, 1996; Enriquez-Obregon et al., 1998; Manickavasagam et al., 2004) (Table 1).
The use of the above genes, in addition to selection of transformed cells and plants, has been beneficial in generating information about transgene expression and stability in transgenic sugarcane (Gallo-Meagher and Irvine, 1996; Enriquez-Obregon et al., 1998; Leibbrandt and Snyman, 2003).
There is limited published work on the use of positive selection systems, such as the E. coli manA phosphomannose isomerase (PMI) gene, in sugarcane. The ubiquitous plant enzyme hexokinase converts mannose to mannose-6-phosphate (Man-6-P). Man-6-P is toxic to plants, but most plants lack PMI and are inhibited by the accumulation of Man-6-P. PMI catalyzes the reversible interconversion of Man-6-P and fructose-6-phosphate, thereby releasing the Man-6-P inhibition and making mannose available as a carbon source for the plant. This system was first demonstrated for transformation of potato, sugar beet, and maize (Bojsen et al., 1998, 1999) and since then it has been used successfully in a variety of other plant species. PMI has no adverse effects in acute mouse oral-toxicity tests, generates no detectable biochemical changes in mannose-associated pathways (Privalle et al., 1999), lacks many attributes known to be associated with allergens (Privalle, 2002), and may thus be considered as an ideal selection protein for plant transformation. For sugarcane transformation, calli were selected on media containing 3gl–1 mannose, in addition to the 20gl–1 sucrose present in the media (Jain et al., 2006). Plant regeneration was performed under the same level of selection. An increase of mannose from 1.5 to 3gl–1 for rooting improved the overall transformation efficiency. The PMI encoding gene, manA, was stably integrated and expressed in almost all of the transgenic lines.
Of the other nonantibiotic systems tested in sugarcane, glutamate-1-semialdehyde aminotransferase was unsuitable, while selection incorporating arabitol showed up to 100-fold lower transformation efficiency. In addition, arabitol is expensive, making it a less desirable selection agent.
2.1.4 Promoters and termination sequences
The two main sources for promoters are from microorganisms (viral or bacterial) or from plants. In addition, there are a few synthetic promoters that have been constructed in the laboratory by combining regulatory and/or enhancer elements from different viral or plant promoters. What is lacking in sugarcane, and in monocots in general, is a promoter that functions as strongly as the CaMV 35S promoter does in dicots. Schledzewski and Mendel 1994 compared transient reporter gene expression in transgenic cells of barley, maize, and tobacco driven by maize polyubiquitin1 (Ubi-1), rice actin1, Emu, or CaMV 35S promoter. CaMV 35S promoter had the highest GUS activity in tobacco (316.71nmolh–1) compared to maize (9.88nmolh–1) and barley (1.22nmolh–1). In contrast, of the four promoters tested, the Ubi-1 promoter showed highest expression in both maize and barley cells, but not in tobacco. However, CaMV 35S in tobacco showed a promoter strength equivalent to 2.5- to 5-fold greater than Ubi-1 in maize (Table 2).
Promoter |
Reporter gene |
Transient expressiona |
Stable expressiona |
References |
|||||
|---|---|---|---|---|---|---|---|---|---|
GUS |
Transient in leaf (after 48h) (nmol 4-MUmin−1mg−1 protein) |
Gallo-Meagher and Irvine, 1993 |
|||||||
Maize Ubi-1 Rice Act1 Emu CaMV 35S |
50.0 (1.00) 10.0 (0.20) 11.0 (0.20) 0.5 (0.01) |
||||||||
GUS |
Percentage of total foci |
Tang et al., 1996 |
|||||||
Total |
BS |
Meso |
Epi |
||||||
Maize Ubi-1 |
771 |
2% |
3% |
95% |
|||||
ScRbcs |
259 |
23% |
8% |
69% |
|||||
Luciferase |
(Fg lucμg−1 protein) 5–8-month-old plants Mean |
Hansom et al., 1999 |
|||||||
Maize Ubi1 Act1 Osa |
84000 (1.00) 1000 (0.12) 3000 (0.04) |
||||||||
Winter 96 |
Summer 96 |
Winter 97 |
|||||||
Maize Ubi-1 |
175 (1.00) |
125 (1.00) |
4500 (1.00) |
||||||
Osa |
25 (0.14) |
10 (0.10) |
70 (0.05) |
||||||
Emu |
5 (0.03) |
2 (0.02) |
200 (0.40) |
||||||
GFP |
Mature Leaf (ugmg−1 protein) Mean |
Schenk et al., 2001 |
|||||||
Maize Ubi-1 BSVCv |
1.5 (1) 4.5 (3) |
||||||||
GUS |
Transient (after 48h) (nmol 4-MUmin−1mg−1 protein) |
Stable (after 4 months) (nmol 4-MUmin−1mg−1 protein) |
Liu et al., 2003 |
||||||
Callus |
Leaf |
Callus |
Leaf |
||||||
Maize Ubi-1 |
1850 (1.00) |
613 (1.00) |
105 (1.000) |
48 (1.0) |
|||||
CaMV 35S |
62 (0.03) |
22 (0.04) |
5 (0.007) |
0 |
|||||
RiceUbiQ2 |
1231 (0.70) |
342 (0.60) |
70 (0.700) |
79 (1.6) |
|||||
GUS |
Transient in callus (nmol 4-MUmin−1mg−1 protein) |
Yang et al., 2003 |
|||||||
Maize Ubi-1 SEF1 SPRP SPRP2.4 |
0.6 0.8 0.9 0.5 |
||||||||
NPTII |
In vitro tissue culture plants (ng NptIImg−1 protein) |
Glasshouse plants (ng NptIImg−1 protein) |
Braithwaite et al., 2004 |
||||||
Callus |
Leaf |
Root |
Meristem |
Leaf |
Root |
||||
Maize Ubi-1 |
100 (1.0) |
50 (1.0) |
75 (1.0) |
45 (1) |
50 (1) |
50 (1) |
|||
SCBV (IMPs) |
100 (1.0) |
85 (1.7) |
80 (1.1) |
225 (5) |
240 (5) |
150 (3) |
|||
SCBV (IMHS) |
130 (1.3) |
100 (2.0) |
60 (0.8) |
250 (6) |
350 (7) |
180 (4) |
|||
GUS |
Stable callus (ng GUSmg−1 protein) Mean |
Wei et al., 2003 |
|||||||
Maize Ubi-1 ScUbi4b ScUbi9b |
1500 (1.0) 600 (0.4) 2000 (1.3) |
||||||||
- a Numbers in brackets are expression relative to that with the Maize Ubi-1 promoter
- b Sc—Sugarcane
2.1.4.1 Viral promoters
The viral promoter most frequently used in plant transformation is the CaMV 35S promoter. However, this promoter is not highly expressed in sugarcane (Table 2). Schenk et al. 2001 reported the isolation of two novel promoters from a DNA virus that infects banana (banana streak virus, BSV). The region of two BSV isolates (2105bp and 1322bp) upstream of the open reading frame (ORF) were labeled as Mys and Cav, respectively, and assessed for promoter activity in stably transformed sugarcane callus using GFP (sGFPS65T) as a reporter gene. Results of the promoter analysis showed that after 19 months growth, the plants transformed with the BSV-Cav construct showed greater than threefold higher activities than the maize Ubi-1 promoter (10.62μgGFPmg–1 total protein and 3.2μgGFPmg–1 total protein, respectively). In addition, this promoter appeared to be expressing in a constitutive manner. GFP accumulation was observed in vascular tissue, bundle sheath cells as well as leaf parenchyma and epidermal cells of sugarcane. Expression, however, was strongest in the parenchyma cells with all three promoters.
Promoters from a sugarcane-specific badnavirus (sugarcane bacilliform virus, SCBV) were cloned and tested using two marker genes (GUS or nptII) (Braithwaite et al., 2004). Three of the four promoters tested were amplified by polymerase chain reaction (PCR) from sugarcane plants containing the virus while the fourth promoter was subcloned from an almost genome-length clone of SCBV. All four promoters were active in sugarcane, with the highest GUS expression present when the subcloned region of SCBV was used for promoter construction. When different parts of the plantlets were assessed for GUS expression (Table 2), the meristems of young plants had the highest levels, there was some in young leaves, but no expression in roots. When nptII expression was assessed in young in vitro plantlets, however, transgene activity was present in callus, leaves, and roots to a similar level (<0.01% of total soluble protein). Thus, the lack of GUS activity in the roots may be a consequence of poor penetration of the GUS substrates to the root region. Interestingly, when the nptII transgenics were grown to maturity in the glasshouse, the lines driven by the SCBV promoter showed a fivefold higher activity than the Ubi-1-driven plants (Table 2).
2.1.4.2 Plant-derived promoters
The maize Ubi-1 promoter (Christensen and Quail, 1996) is the most frequently used constitutive promoter for sugarcane transformation (Table 2) and its expression is usually higher than other plant-derived promoters (sugarcane Ubi and rice actin1). Expression of luc activity in sugarcane plants by maize Ubi-1 promoter ranged between 200 and 300000 relative light units (RLU) mg–1 protein compared to 200–2000 RLUmg–1 protein in actin1 lines (Hansom et al., 1999; Table 2). In addition, this expression was independent of copy numbers.
Using particle bombardment of sugarcane callus, Liu et al. 2003 showed that there was a 1.6-fold increase in GUS expression by the rice Ubi-2 promoter (mean = 78.6nmol 4-MUmin−1mg−1 protein) over the maize Ubi-1 promoter (mean = 47.6nmol 4-MUmin−1mg−1 protein). The GUS gene driven by CaMV 35S showed no expression in regenerated plants, and was <10% of that of ubiquitin promoters in transient expression analysis (Table 2).
Tang et al. 1996 compared the expression patterns of GUS, driven by promoters of two sugarcane ribulose-1,5-bisphosphate small subunit (Rubisco) genes (scrbcs-1 and -2), in transient assays. Both promoters directed expression in leaf tissues, but not in calli. Although the overall expression levels were lower when compared to the maize Ubi-1 promoter, scrbcs promoters drove higher expression in the photosynthetic cells, especially in the bundle sheath cells (Table 2). Stable expression in transgenic callus lines and regenerated plants also showed that scrbcs-1 promoter directed GUS expression in leaves, but not in calli.
Two sugarcane polyubiquitin gene promoters (sugarcane ubi4 and ubi9) were found to direct high levels of transient GUS expression in the following monocots: sugarcane, maize, sorghum, banana, pineapple, garlic, and rice cells (Wei et al., 1999, 2003). Both of these monocot promoters were also sufficient to drive GUS expression in cells of tobacco. In these transient assays, the activities of the two sugarcane promoters were comparable to the strong monocot promoter, maize Ubi-1 (Christensen and Quail, 1996). Similar to Ubi-1, sugarcane ubi4 was heat shock inducible in stably transformed sugarcane callus lines, but sugarcane ubi9 was not (Wei et al., 2003). The physiological difference between the two sugarcane ubiquitin promoters corresponded to a MITE (miniature inverted-repeat transposable element) insertion that is present in the putative heat shock elements of sugarcane ubi9 but not present in sugarcane ubi4. In transgenic sugarcane plants produced by particle gun bombardment, GUS expression from sugarcane ubi4 and ubi9 dropped to very low or nondetectable levels after plant regeneration. This drop in expression also occurred in Ubi-1 sugarcane lines. Nuclear run-on experiments showed the down-regulated transgenes continued to be transcribed at high levels, indicating that the lack of transgene expression was due to post-transcriptional gene silencing (PTGS). In contrast, sugarcane ubi9 drove high levels of expression in transgenic rice plants produced via Agrobacterium-mediated transformation. This high level of expression continued after plant regeneration and was inherited in the T1 generation (Wei et al., 2003).
2.1.4.3 Synthetic promoters
The promoter Emu was synthesized by Last et al. 1991 and contains a truncated maize alcohol dehydrogenase (Adh) promoter as well as several copies of the maize anaerobic responsive elements from the Adh gene. In addition, it contains the ocs-element of the octopine synthase gene from Agrobacterium. Comparative studies of GUS expression in sugarcane protoplasts using constructs with either the Emu or the CaMV 35S promoter showed a 50–100-fold increase in Emu-driven GUS expression over the 35S promoter (Rathus and Birch, 1992). However, Joyce et al. 1998 and Bower et al. 1996 found that the Emu promoter could not drive strong expression in mature sugarcane plants (Table 2).
Osa is another synthetic promoter, developed by CSIRO Plant Industry Australia, which consists of multiple octopine synthase (OCS) enhancer elements, the core region from the CaMV 35S promoter and untranslated leader sequence from maize transposable element Ac (Bower et al., 1996). Experiments in sugarcane using this promoter also showed much lower levels of gene expression than maize Ubi-1 (Table 2).
2.1.4.4 Terminator sequences
The most commonly used terminator sequences for transformation of sugarcane callus is the Nos terminator sequence from Agrobacterium. There are a few reports where the Agrobacterium octopine synthase gene (Ocs) terminator sequence has been used (Elliott et al., 1998). The studies have not compared the effect of terminator sequences on transgene expression in sugarcane; this is an area of research that has been neglected and warrants further investigation.
2.1.5 Activity, stability of inheritance, and silencing of transgenes
The high level of ploidy found in all sugarcane may have some important implications for transformation, which are not normally encountered when working with diploid species. For instance, increase in ploidy by genome duplication, which has been suggested to play an important role in angiosperm evolution (Stephens, 1951; Ohno, 1970; Blanc et al., 2000; Initiative, the Arabidopsis Genome, 2000; Paterson et al., 2000, 2003, 2004, 2005; Bowers et al., 2003; Vandepoele et al., 2003; Wang et al., 2005b), is associated with a host of rapid responses, including loss and restructuring of low-copy DNA sequences (Song et al., 1995; Feldman et al., 1997; Ozkan et al., 2001; Shaked et al., 2001; Kashkush et al., 2002), activation of genes and retrotransposons (O'Neill et al., 1998; Kashkush et al., 2003), gene silencing (Chen and Pikaard, 1997a, b; Comai, 2000; Comai et al., 2000; Lee and Chen, 2001), and organ-specific subfunctionalization of gene expression patterns (Adams et al., 2003, 2004). Sugarcane appears to have undergone two such genome duplications in about the past 5 million years and these mechanisms may be especially important in providing raw material for evolutionary change. It is important to understand how stable the sugarcane genome is with respect to: (i) transposon activity; (ii) genome expansion or contraction; (iii) measuring the extent of gene silencing and alterations in gene expression; (iv) assessing in vitro regeneration systems for epigenetic variation in regenerated plantlets; and (v) designing transformation technologies to overcome the above constraints.
Molecular techniques for detection and characterization of transgenes with respect to integration pattern, copy number, and protein concentrations have been conducted and published (Bower et al., 1996; Gallo-Meagher and Irvine, 1996). However, these analyses were carried out on sugarcane plants maintained in glasshouses. Field analysis to determine expression and stability of transgenes is important because of sugarcane's multiple vegetative crop cycles and reports of PTGS.
The transgenesis approach in sugarcane has targeted elite commercial cultivars. It would be advantageous to be able to use transgenic plants as parents in breeding programs especially when the desired trait is not present in the sugarcane gene pool. Transgene inheritance and segregation of the bar herbicide resistance gene and the hut Sorghum mosaic virus (SrMV) coat protein gene was tracked in progeny arising from conventional crosses made between transgenic and nontransgenic parents (Butterfield et al., 2002). The results demonstrated that transgenic plants can be used as parents in a sugarcane breeding program but screening progeny for a characteristic such as virus resistance that relies on PTGS mechanisms may have to be carried out after one vegetative cycle of growth after crossing.
PTGS is known to occur in sugarcane (Ingelbrecht et al., 1999) and is considered to be one of the factors limiting accumulation of recombinant proteins (Wei et al., 2003). It was thought that it could be reversed by retransformation with a gene encoding a viral suppressor of PTGS, such as HcPro from SrMV (Ingelbrecht et al., 2000) and P0 from sugarcane yellow leaf virus (ScYLV) (Wang et al., 2006, 2007). Despite the potential application in transgene silencing control, there are possible negative effects on endogenous gene regulation and virus susceptibility. Initial results indicate that there is no consistent correlation between the RNA expression levels of P0 or HcPro and the expression of a transgene and several miRNA-regulated endogenous genes. Further research is required to characterize the effect of these viral suppressors in transgenic sugarcane (Wang et al., 2006, 2007).
Sugarcane plants transformed via particle bombardment with GM-CSF (human cytokine granulocyte macrophage colony stimulating factor), contain numerous copies of transgenes with complex integration patterns as revealed by Southern blot hybridization (Albert et al., 2003; Wang et al., 2003). Multiple approaches have been evaluated for the introduction of single- or low-copy transgenes, to determine if these methods can reduce transgene silencing (Albert et al., 2004). These methods include the use of insert-only DNA for bombardment (Fu et al., 2000), Cre/lox site-specific recombination to resolve multiple transgene copies (Srivastava and Ow, 2001), and the Ac/Ds transposon system to direct transgene integration by transposition (Koprek et al., 2000, 2001). The use of linear expression cassette-only DNA was reported to produce a high frequency of low-copy transgene insertions in rice, with no evidence of silencing through the R4 generation (Fu et al., 2000). In contrast, most transgenic sugarcane lines produced by this method contained multiple transgene copies, with only three of 27 selected lines containing three or fewer copies (Wang et al., 2003). Cre/lox lines did contain fewer copies of the transgene as estimated by quantitative PCR (qPCR). However, accumulation of GM-CSF in the low-copy lines was not higher than in multicopy lines (Albert et al., 2004). In addition, a vector system was developed to allow direct plasmid to chromosome transposition using Ac/Ds in monocot cells. This should allow single-copy transposition lines to be produced in a single generation without sexual crosses (Albert et al., 2003). However, no increase in GM-CSF protein level was observed in the Ac/Ds lines.
2.1.6 Adverse effects on growth, yield, and quality
Most evaluation of transgene stability and expression patterns and performance of transgenic sugarcane plants in the field has been limited to a few lines (Gallo-Meagher and Irvine, 1996; Arencibia et al., 1999; Leibbrandt and Snyman, 2003). However, results of recent studies where a larger number of lines were tested (Gilbert et al., 2005; Vickers et al., 2005b) may influence future transgenic approaches.
Initial field trials demonstrated stable expression of a herbicide-resistant transgene over three rounds of vegetative propagation in a single transformant (Gallo-Meagher and Irvine, 1996; Leibbrandt and Snyman, 2003), but no agronomic measurements were taken in the first study. In the latter trial, no differences were found between the transgenic line and wild-type control in phenotypic characters such as stalk height and diameter, agronomic performance indicators such as sucrose yield and fiber content, and disease susceptibility ratings to smut and rust. However, in a field trial where 100 transgenic lines were compared for Sugarcane mosaic virus (SCMV) resistance and yield characteristics, a considerable amount of variability for measured parameters was reported, which the authors attributed largely to the effects of the cultivar used and the tissue culture process (Gilbert et al., 2005; Vickers et al., 2005b). Similarly, preliminary data from metabolomic analysis, comparing leaves from nontransformed sugarcane with leaves from transgenic sugarcane lines producing polyhydroxybutyrate (PHB), found that the vast majority of the variation was a tissue culture effect and was not from the insertion of the PHB metabolic pathway and the selectable marker genes (Purnell et al., 2007).
2.1.7 Subcellular targeting
Most transgene expression constructs used for sugarcane transformation result in production of foreign gene products in the cytosol, but important metabolic activities ranging from photosynthesis to sugar storage are carried out in other compartments. Hence, targeting proteins to subcellular compartments may be necessary for effective resistance to pest and diseases and efficient metabolic engineering in sugarcane. For example, for increased resistance to sugarcane leaf-scald disease, resistance gene products may need to be targeted to the plastids, as the albicidin toxin from the pathogen Xanthomonas albilineans blocks plastid DNA replication and chloroplast development. Similarly, for efficient conversion of sucrose to alternative carbohydrates such as starch and fructans, foreign proteins need to be targeted to the sugarcane vacuoles in mature stem parenchyma cells, where most of the sucrose is stored.
The ability to target recombinant proteins to the correct subcellular location using efficient and appropriate targeting signals is one of the most important requirements for sugarcane metabolic engineering. Various targeting signals have been successfully tested to target heterologous proteins to different subcellular compartments in sugarcane. The availability of visible reporters such as GFP in combination with a transient assay system in sugarcane leaves allowed the testing of the efficiency of various targeting signals with less time and resources (Gnanasambandam et al., 2007). While most of the tested signal sequences are from dicotyledons species, they were effective in the monocotyledon sugarcane. So far, targeting signals for vacuoles, endoplasmic reticulum (ER), plastids, mitochondria, and peroxisomes have been tested in sugarcane (McQualter et al., 2005; Petrasovits et al., 2007; Brumbley et al., 2006b; Gnanasambandam et al., 2007). In future, signals for other compartments and efficiency of signals will be determined.
2.1.7.1 Protein targeting to vacuoles
Sugar storage vacuoles occupy about 80% of the total tissue space in mature sugarcane stem and accumulate sucrose up to 500μmolg–1 fresh weight (Moore, 1995). Due to the potential to engineer synthesis of valuable compounds other than sucrose in sugarcane (e.g., alternative carbohydrates such as starch and fructans), the sugarcane vacuole is one of the important target compartments for metabolic engineering. However, targeting proteins to the sugarcane vacuoles remains a challenge and differs in several aspects compared to targeting to other compartments: (i) the sugarcane vacuolar compartment is highly dynamic in function, and is acidic as indicated by neutral red accumulation in most protoplasts isolated from sugarcane suspension cells and stem storage parenchyma cells (Gnanasambandam and Birch, 2004). It is not known whether several vacuole types coexist in sugarcane cells with different pH and/or proteolytic activities. As a result, there may be several independent targeting mechanisms to different vacuole types in sugarcane; (ii) targeted proteins should be stable and functional in the vacuolar environment, engineered for the relevant pH and proteases. For example, when the N-terminal vacuolar targeting signal (NTPP) of potato patatin was used to target yeast invertase to the sugarcane vacuole, neither detectable amounts of invertase protein nor increased soluble acid invertase activity was observed (Ma et al., 2000). Similarly, fusion of NTPP from sweet potato sporamin to various reporter proteins resulted in substantial reduction or loss of enzymatic activity in transient expression assays and in transformed sugarcane cells (Gnanasambandam and Birch, 2004); and (iii) protein targeting mechanisms to many compartments (e.g., peroxisomes, mitochondria, and ER) are highly conserved in eukaryotes. In contrast, protein targeting to the plant vacuole through the endomembrane system is different from the lysosomal and vacuolar targeting mechanisms of animals and yeast. While mammalian cells use mannose-6-phosphate mediated lysosomal targeting, yeast cells use N-terminal propeptides for vacuolar targeting. In contrast, plants employ three different types of targeting signals that are found either at the N-terminus or the C-terminus or within the mature polypeptide (Matsuoka and Neuhaus, 1999).
Two signals, the NTPP of sweet potato sporamin and sugarcane legumain, were reported to be efficient in targeting reporter proteins to the sugarcane vacuole (Gnanasambandam and Birch, 2004; Rae et al., 2006). In contrast, the C-terminal signal from tobacco chitinase was inefficient in targeting a reporter protein to the sugarcane vacuole (Gnanasambandam and Birch, 2004). Using the NTPP of sporamin, metabolic engineering of vacuolar compartment to produce high-value isomaltulose has been demonstrated successfully (Wu and Birch, 2007). Vacuolar targeting of the sucrose isomerase using the NTPP of sporamin allowed high isomaltulose yields (up to 440μmolg–1 fresh weight) in sugarcane stems. In contrast, expression of a cytosolic form of the same enzyme caused stunting with reduced sugar accumulation. Interestingly, isomaltulose accumulated in storage tissues without any decrease in stored sucrose concentration, resulting in doubled total sugar concentrations in harvested juice (Wu and Birch, 2007) in vacuolar targeted lines. The transgenic lines with enhanced sugar accumulation also showed increased photosynthesis, sucrose transport and sink strength (Wu and Birch, 2007).
2.1.7.2 Protein targeting to plastids
The N-terminal plastid transit peptides of RbcS (rubisco small subunit) genes from maize and pea were used to target heterologous enzymes to sugarcane plastids to successfully produce p-hydrobenzoic acid (pHBA; McQualter et al., 2005) and polyhydroxybutyric acid (PHB biopolymer; Petrasovits et al., 2007). The transit peptides from tomato DCL (defective chloroplast and leaves) and tobacco RbcS genes were shown to target GFP to the sugarcane leaf proplastids (Gnanasambandam et al., 2007; Figure 4).
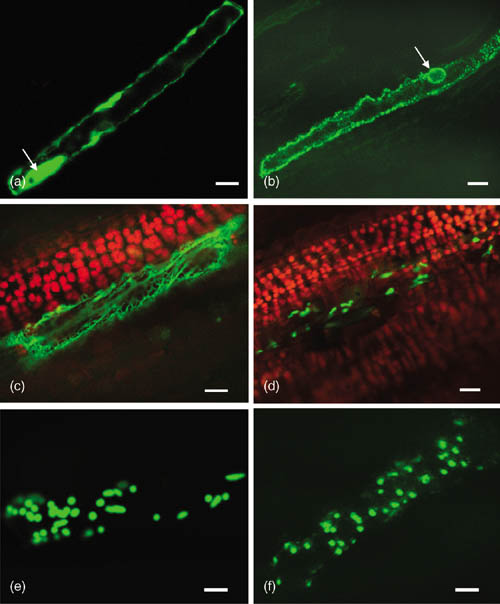
Confocal images of sugarcane leaf epidermal cells showing fluorescence of green fluorescent protein (GFP) in different subcellular compartments. Nontargeted GFP in the cytosol (a) and nucleus (arrow in a). ER-targeted GFP fluorescence in the ER (b, c; arrow in b shows perinuclear distribution). Plastid-, mitochondrial-, and peroxisomal-targeted GFP in the proplastids (d), mitochondria (e), and peroxisomes (f), respectively. All images show green channel GFP fluorescence except (c) and (d) that show merged images of green (GFP fluorescence) and red (chlorophyll autofluorescence) channels. Bar = 10μM [Reproduced with permission from Annathurai Gnanasambandam, BSES Limited, Australia]
2.1.7.3 Protein targeting to mitochondria
Mitochondria are important organelles involved in ATP synthesis, photorespiration, and programmed cell death. The N-terminal mitochondrial presequence from F1-ATPase β-subunit (ATPase-β) of Nicotiana plumbaginifolia was shown to be effective in targeting GFP to the mitochondria (Petrasovits et al., 2007; Figure 4). Although this signal was used to target bacterial enzymes to sugarcane mitochondria, no PHB polymer accumulation was observed in transgenic sugarcane (Petrasovits et al., 2007).
2.1.7.4 Protein targeting to peroxisomes
Plant peroxisomes are involved in fatty acid β-oxidation, the glyoxylate cycle and photorespiration. The six amino acid C-terminal peroxisomal signal from Spinacia oleracea L. (spinach) glycolate oxidase was shown to be effective in targeting GFP to the peroxisomes in sugarcane leaves and callus (Gnanasambandam et al., 2008; Figure 4).
2.1.7.5 Protein targeting to ER
Almost all of the proteins that will be secreted to the cell exterior, as well as those destined for the lumen of the ER, Golgi apparatus, or vacuoles, are initially delivered to the ER lumen (Gal, 1998). The N-terminal ER signal peptide from Arabidopsis basic chitinase and a C-terminal HDEL signal for protein retention in the ER was efficient in targeting and retention of GFP in the ER in sugarcane leaves (Gnanasambandam et al., 2007; Figure 4). This HDEL signal in combination with the ER signal peptide of PinII was required for higher accumulation of GM-CSF in transgenic sugarcane (Wang et al., 2005a).
2.2 Target Traits and Products
For sugarcane genetic engineering, specific targets fall into two broad areas: (i) input traits that improve crop performance and productivity such as pest and disease resistance, tolerance to abiotic stress, herbicide tolerance, and alterations to plant architecture and (ii) output traits that modify quality and yield, compositions and use, such as production of more sucrose, biomass or novel compounds.
2.2.1 Disease resistance
Reports of pathogen-derived resistance to virus diseases include resistance to SCMV in otherwise susceptible sugarcane clones (Joyce et al., 1998; Ingelbrecht et al., 1999). Both groups used the coat protein gene of the virus driven by the Ubi-1 promoter with the nos terminator. Plants transformed with the coat protein gene of the SrMV strain SCH displayed a range of phenotypes, including immune, resistant, recovery, and susceptible plants, when challenged with virus. These observations enabled insights to the RNA-mediated PTGS resistance mechanism (Ingelbrecht et al., 1999). Virus induced gene silencing (VIGS) has been suggested as the mechanism of resistance against viral infections. Interestingly, transgenic plants having the same transgene integration pattern as determined by Southern blot analysis (i.e., clones) displayed a different response to mechanical SCMV infection. The reason for this peculiar result is still not clear (Ingelbrecht et al., 1999).
Sugarcane yellow leaf disease, which is characterized by yellowing of the leaf midribs followed by tissue necrosis, is caused by ScYLV (Borth et al., 1994; Schenck et al., 1997; Vega et al., 1997; Comstock et al., 1998). Economic losses from sugarcane yellow leaf disease of up to 50% have been reported (Vega et al., 1997). The ScYLV coat protein gene in the sense orientation driven by Ubi-1 was used to generate virus-resistant transgenic sugarcane. Resistance levels were evaluated with inoculation of viruliferous aphids. Virus titers were determined using tissue blotting and quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR). Two resistant transgenic sugarcane lines were identified based on the tissue blot analyses. However, virus RNA can still be detected using qRT-PCR in these lines. Greenhouse tests are currently being conducted to compare the yield difference between nontransformed and transgenic lines.
The transgenic approach for providing resistance to the more devastating viral disease in sugarcane, Fiji leaf gall has been tested (McQualter et al., 2004). Resistance to Fiji leaf gall was produced by microprojectile-mediated transformation with a transgene encoding a translatable version of Fiji disease virus (FDV) segment 9 ORF 1 under the control of the maize Ubi-1 promoter. The molecular phenotypes of the transgenic plants at both the DNA and RNA levels were not entirely consistent with a resistance mechanism based on PTGS. Transgenic plants showed very low steady state messenger-RNA (mRNA) levels under normal conditions, but many of these plants failed to show resistance upon challenge with the FDV virus, suggesting that the virus possessed a mechanism for overriding the post-transcriptional silencing mechanism. Further research required to achieve complete immunity to FDV includes: (i) additional characterization of the FDV genome, since only a superficial knowledge of the virus life cycle and replication strategy is currently known; (ii) a shotgun approach where all 12 ORFs contained within the FDV genome are used as transgenes, either singly or in various combinations (RNAi (RNA interference) silencing constructs should be employed in this case); and (iii) determining if FDV possesses a dsRNA (double-stranded RNA) binding protein with the ability to suppress gene silencing. If so, then a more effective approach to achieve pathogen derived resistance to FDV in sugarcane might require deactivation of this protein.
Leaf scald is a serious disease of sugarcane caused by the bacterium Xanthomonas albilineans. The bacterium produces a toxin (albicidin) that blocks plastid DNA replication of the sugarcane plant. Zhang et al. 1999 identified a bacterium that could survive in the presence of albicidin because it carried an albicidin detoxification gene (albD). This gene was cloned from the bacterium and introduced into sugarcane. Some of the resulting transgenic plants were resistant to leaf scald disease (Hansom et al., 1999). It is interesting to note that another bacterial pathogen of sugarcane Leifsonia xyli subsp. xyli (Lxx) like Xanthomonas albilineans colonizes the xylem vessels, was recently shown to have a gene homologous to the one in Xanthomonas albilineans encoding an enzyme to pump albicidin out of its cells (Monteiro-Vitorello et al., 2004).
2.2.2 Insect resistance
The potential for using a modified Bt gene encoding the δ-endotoxin CryIA(c) from Bacillus thuringiensis strain kurstaki to combat damage by the lesser corn-stalk borer (LCB) was demonstrated by Fitch et al. 1996. The Bt gene and the selectable marker gene nptII, were under the control of the CaMV 35S promoter. Bombarded sugarcane calli were selected stepwise on 50–200mgl–1 of G418, an aminoglycoside antibiotic similar in structure to gentamicin B1. Insect bioassays indicated that LCB larvae fed on some lines of calli or leaves from the regenerated plants weighed less and showed higher mortality than those fed on nontransgenic tissues. Mortality was higher when LCB were fed transgenic calli as opposed to transgenic leaves, possibly due to different levels of the CryIA(c) protein in the two different tissues, although the protein levels were not reported. Leaves from other putatively transformed lines had no detectable effect on larvae survival. Southern hybridization indicated that there were 1–5 copies of the Bt genes in the two most resistant lines.
Resistance to the other lepidopteran sugarcane stalk borers has also been reported using the δ-endotoxin gene from B. thuringiensis. Larval mortality and reduced levels of damage from Diatraea saccharalis in the field and Proceras venosatus in the glasshouse have been reported (Arencibia et al., 1997; Weng et al., 2006). In the short term, it is possible that intellectual property restrictions may limit widespread use of this technology in sugarcane. Consequently, the use of other antimetabolic compounds for improving plant resistance, such as proteinase inhibitors (Allsopp et al., 1996; Falco and Silva-Filho, 2003) and lectins (Sétamou et al., 2002a, 2002b, 2002c), has been tested with promising results from laboratory-based insect bioassays. These products impact on a wider range of insect/pests and studies on the effects on the Australian coleopteran whitegrub have been underway for several years (Allsopp et al., 1996).
Canegrubs are a major pest in the Australian sugar industry causing yield losses up to AU $80 million. The PinII and the snowdrop lectin (Gna) genes have both been used to generate transgenic sugarcane in attempts to control these pests (Allsopp et al., 1996; Nutt et al., 1999). Plants containing the pinII gene grew more slowly than the nontransformed control plants. This may have been due to metabolic disruption within the plants, as no pinII was found in the cell vacuole. Earlier work with artificial feeding trials had shown that avidin could reduce larval growth and increase larval mortality of Antitrogus parvulus (Childers canegrub) (Allsopp and McGhie, 1996). More recently, the gene for avidin, a biotin binding protein from chicken egg white, has been introduced into sugarcane for the control of canegrubs (Nutt et al., 2006). Transgenic sugarcane plants have been regenerated, which contain avidin concentrations of up to 0.06% of total protein.
2.2.3 Sucrose metabolism
A suite of physiological processes and enzymes involved in sucrose accumulation have been identified and characterized over the last 50 years. These processes include: leaf reactions, such as photosynthetic reactions, sucrose synthesis, metabolism and carbon partitioning across various membranes into different pools; phloem reactions, such as phloem loading in leaf, translocation to and unloading in various sink tissues (including primary storage in parenchyma cells of the stalk); stalk reactions, such as membrane transport, sucrose metabolism, carbon partitioning, and remobilization of stored sucrose; genetic and developmental controls, such as timing of maturation; and environmental perception and signal transduction pathways to coordinate plant development (Moore, 2005). Some of the genes encoding these enzymes have been cloned and used to transform sugarcane with the goal of altering sucrose accumulation (reviewed by Grof and Campbell, 2001). However, this reductionist approach has fallen short of expectation in almost all of the cases because of the complexity among the multitude of simultaneous processes and parallel pathways.
As tissue culture methods became established, they were exploited to elucidate the physiology and biochemistry of sugarcane carbohydrate accumulation, with particular emphasis on sugars (Komor et al., 1981; Thom and Komor, 1984). Transgenic sugarcane cell lines were also used to study the effect of invertase expression in different cellular compartments on sucrose accumulation (Ma et al., 2000). Overexpression of a yeast invertase gene (SUC2) in the apoplast led to rapid hydrolysis of sucrose and accumulation of hexoses, both in the medium and the cells, suggesting that hexose uptake, not hexose availability, was the limiting factor for sucrose accumulation. Cells transformed for overexpression of invertase in the cytoplasm did not show a significant change in the sugar composition in the medium, but did significantly reduce the sucrose content in the cells. Partial inhibition of the soluble invertase activity was achieved by transforming with a sugarcane soluble acid invertase complementary DNA (cDNA) (SCINVm) in the antisense orientation to result in increased sucrose accumulation. Intra- and extra-cellular sugar composition was very sensitive to changes in invertase activities in this tissue culture system (Ma et al., 2000).
Pyrophosphate-dependent phosphofructokinase (PFP) activity in sugarcane is inversely correlated to sucrose concentration in maturing internodal tissues, but no clear physiological role in sucrose metabolism has emerged (Whittaker and Botha, 1999). If endogenous PFP activity were to be down-regulated by antisense or co-suppression technologies, then sucrose concentration could be increased. In sugarcane transformed with the catalytic subunit, PFP-β, endogenous PFP gene expression was reduced by up to 40% and 80% in leaf roll and internodal tissue, respectively (Groenewald and Botha, 2001, 2008). Sucrose concentrations in these lines were significantly increased in immature internodes. This finding could make a valuable contribution to the productivity of sugarcane cultivars and elucidate the role of PFP in sucrose accumulation.
Vickers et al. 2005a attempted to modify the endogenous polyphenol oxidase (PPO) activity in sugarcane by introducing sense and antisense constructs of the endogenous sugarcane PPO gene driven by the Ubi-1 promoter. The rationale behind this approach was that the inhibition of PPO activity in juice by chemical inhibitors, elevated pH or heat significantly reduced the color of the cane juice and the subsequent color intensity in sugar crystals. All transgenic lines, irrespective of the orientation of the PPO gene, showed higher PPO activity and more color units than the nontransformed commercial clones. Although higher levels of PPO activity were correlated to juice with a darker color, the converse was not true in this study. It has been hypothesized that a lowering of PPO activity will lead to a reduction in the color of juice and consequently raw sugar (Vickers et al., 2005a). As pale sugar has a market premium over dark sugar, this has potential economic benefits for the sugar industry.
While most transgenic attempts to improve sugar accumulation in sugarcane have met with limited success, a notable exception was the introduction of a bacterial gene encoding a sucrose isomerase (Wu and Birch, 2007) to produce a high-value sugar isomaltulose.
2.2.4 Alternative products
High-value sugars. Isomaltulose is a sucrose isomer with α-1,6 linkages instead of α-1,2. Although it is only 42% as sweet as sucrose (Li et al., 2004), it is a high-value sugar because it is digested 4–5 times slower than sucrose and so has major health benefits to consumers (Lina et al., 2002). Not only does isomaltulose reduce the highs and lows in blood sugar (Lina et al., 2002), it also has dental health benefits (Ooshima et al., 1983). Wu and Birch 2007 demonstrated that sugarcane can be engineered to target the production of isomaltulose in the storage vacuoles in sugarcane stalks (see Section 2.1.7). In transgenic lines, when sucrose isomerase was targeted to the vacuole, isomaltulose accumulated without a reduction in sucrose accumulation. Up to a twofold increase in total sugar accumulation was observed in some lines (Wu and Birch, 2007). The additional sugar was not produced at the expense of structural carbohydrates, as levels of fiber were the same between wild type and transgenic lines. Additional benefits were that the levels of isomaltulose reduced leaf senescence resulting in more leaf biomass on the plants and a reduction in the percent water in stalk tissue from 70% for the nontransformed control mature stalk internode to 60% in the best isomaltulose producing transgenic line (Wu and Birch, 2007). Field trials of these high-sugar lines are currently underway.
Sorbitol. The primary photosynthate of the members of the Rosaceae family including apples (Malus domestica), pears (Pyrus sp.), peaches and nectarines (Prunus persica), plums (Prunus subgenus Prunus), cherries (Prunus subgenus Cerasus) is sorbitol. Sorbitol has intrinsic value as a noncaloric sweetener and is also used to manufacture ascorbic acid and personal care products (Kirschner, 2004). Sorbitol synthesis in sugarcane is achieved by a single enzyme conversion step sorbitol-6-phosphate dehydrogenase (S6PDH), which catalyzes the reduction of D-glucose-6-phosphate (G6P) to sorbitol-6-phosphate (S6P). The cytosolic expression of the M. domestica sorbitol-6-phosphate dehydrogenase gene (mds6pdh) resulted in high accumulation of sorbitol (up 61% of the soluble sugars or 12% of the leaf dry weight) in sugarcane leaves but with 10-fold less sorbitol in the culm (Chong et al., 2007; Brumbley et al., 2004). Sugarcane leaves developed necrosis in a pattern characteristic of early senescence and the severity was related to the relative quantity of sorbitol accumulated. Vacuolar targeting of the same enzyme, while not attempted, may lead to normal plants and higher accumulation of sorbitol in the culm as observed for isomaltulose (Wu and Birch, 2007). However, protein stability within the vacuole will continue to be a major challenge for efficient metabolic engineering of the sugar storage compartment in sugarcane. A possible solution would be to modify the enzymes, possibly by using directed evolution (Stemmer, 1994a, 1994b; Chica et al., 2005; Kaur and Sharma, 2006) to function optimally at the pH and to be more resistant to the protease activity in the environment of the storage vacuole.
PHB sugarcane line TA4 cells showing plastids containing polyhydroxybutyrate granules. Six-month-old sugarcane plants were sectioned and analyzed for PHB content in (a) leaf tissue taken from the midrib of the lowest green leaf and sampled from older tissue closest to the leaf tip, (b) stem tissue sampled from the basal internode, (c) stem tissue sampled from the basal node. Black and white arrows point to clusters of PHB granules [Reproduced with permission from Todd Werpy and Gene Petersen, the lead authors of “Top Value Added Chemicals from Biomass”. http://www1.eere.energy.gov/biomass/pdfs/35523.pdf]
In addition to the above mentioned sugars, trehalose, a nonreducing disaccharide of glucose, has also been produced in sugarcane (Brumbley et al., 2006b; O'Neill et al., 2006; Zhang et al., 2006). Trehalose may play a role in carbohydrate metabolism impacting on glucose, fructose, and sucrose levels (Rontein et al., 2002).
Biopolymers. To test the ability of sugarcane to be a biofactory, the products from the Ralstonia eutropha PHB biosynthetic pathway were targeted to several subcellular compartments of sugarcane (Brumbley et al., 2002, 2004, 2007; Petrasovits et al., 2007; Purnell et al., 2007). PHB is the best-studied member of the polyhydroxyalkanoate (PHA) family, of which approximately 130 naturally occurring members have been identified (see Section 3). PHB is produced from acetyl-coenzyme A (acetyl-CoA) by the successive action of three enzymes [ketothiolase (PHAA), acetoacetyl-reductase (PHAB), and PHB synthase (PHAC)], which are encoded by the genes phaA, phaB, and phaC, respectively (Peoples and Sinskey, 1989a, 1989b). Each gene was on a separate transformation vector and gene expression was controlled by Ubi-1 promoter and nos terminator sequences. The three vectors with the PHB biosynthetic pathway were biolistically transformed into sugarcane callus simultaneously along with a construct containing the selectable marker gene nptII (Petrasovits, 2005; Petrasovits et al., 2007).
Previous attempts to produce PHB at high levels in plants resulted in either low levels of PHB accumulation or severe negative phenotypic effects. In sugarcane, the polymer accumulated in the leaves of chloroplast-targeted lines at levels up to 2.5% of dry weight and 0.01% in stems (Figure 5) (Petrasovits et al., 2007). Purnell et al. 2007 conducted a replicated glasshouse trial using a random block design with six independent PHB producing transgenic sugarcane lines and found that stalk height and weight and sugar levels were not affected by PHB accumulation.
Sugarcane has also been evaluated as a production platform for p-hydroxybenzoic acid (pHBA) using two different bacterial proteins (a chloroplast-targeted version of E. coli chorismate pyruvate-lyase and a 4-hydroxycinnamoyl-CoA hydratase/lyase from Pseudomonas fluorescens) (McQualter et al., 2005; Brumbley et al., 2004). Both lines provide a one-enzyme pathway from different naturally occurring plant intermediates. The substrates for these enzymes are chorismate (a shikimate-pathway intermediate that is synthesized in plastids) and 4-hydroxycinnamoyl-CoA (a cytosolic phenylpropanoid intermediate). Although both proteins have previously been shown to elevate pHBA levels in plants (Siebert et al., 1996; Mayer et al., 2001), they had never been evaluated concurrently nor had they been used simultaneously in the same plant. The pHBA was quantitatively converted to glucose conjugates by endogenous uridine diphosphate (UDP)-glucosyltransferases and was stored in the vacuole. The largest amounts detected in leaf and stem tissue were 7.3% and 1.5% of dry weight (DW), respectively, and there were no observable phenotypic abnormalities. However, as a result of diverting carbon away from the phenylpropanoid pathway, there was a reduction in leaf chlorogenic acid, subtle changes in lignin composition, and an apparent compensatory up-regulation of phenylalanine ammonia-lyase (PAL; McQualter et al., 2005).
2.2.5 High-value protein production
GM-CSF, which is used in clinical applications for treatment of neutropenia and aplastic anemia, was used to test the feasibility of using sugarcane as a biofactory for high value protein production (Wang et al., 2005a). Two promoters, Ubi-1 and sugarcane ubi9 were tested in transgenic sugarcane lines resulting in production of up to 0.02% total soluble protein as GM-CSF. No significant difference was observed between the Ubi-1 or ubi9 promoter line. To achieve higher accumulation of GM-CSF in sugarcane, a C terminal HDEL tag for ER targeting had to be added to the gene construct. The sugarcane-produced GM-CSF showed identical biological activities when compared to commercially available purified protein. In a 14-month old field trial, no abnormal phenotype was observed and the accumulation levels remained relatively stable. This is the first report of GM-CSF production in field-grown plants. No flowering of the trial plants occurred and no pollen or seed was produced during the trial period. Drying, burning, and burial of the test plants effectively blocked possible routes for the transgenic sugarcane to enter the food supply or environment. If GM-CSF accumulation in sugarcane had been as high as Sétamou et al. 2002c reported for the snowdrop lectin (1–1.25% total soluble protein), commercial production would probably have been economically viable because of the high commercial value of this protein.
Sugarcane has also been used for the production of bulk protein, collagen. In vertebrates, the collagen family is made up of approximately 27 proteins (Myllyharju et al., 2000; Kielty and Grant, 2002; Gelse et al., 2003; Myllyharju and Kivirikko, 2004; Pakkanen et al., 2006). Collagens make up 25% of the total proteins in humans and function to act as connecting structures and to give mechanical stability to the entire body. They form triple-helical structures, giving them commercial importance for the food industry and for a range of medical applications, such as tissue repair and cosmetic surgery (Bulleid et al., 2000). Gelatin is denatured collagen. The yeast Pichia pastoris has been engineered successfully for the high level production of triple-helical collagen (Pakkanen et al., 2006). Production has also been attempted in sugarcane and a fragment of human collagen was produced at very low levels (0.025% total soluble protein) (E. Mirkov, personal communication).
2.2.6 Abiotic stress
One strategy for generating abiotic stress-resistant plants has been to express genes for the production of trehalose. In planta trehalose production confers stress tolerance in plants (Garg et al., 2002; reviewed in Penna, 2003; Rolland et al., 2006). Abiotic stress normally causes a reduction in photosynthesis. However, trehalose protects the photosynthetic machinery in the plant cells and allows them to remain functional for longer periods of time under a range of abiotic stress conditions (Garg et al., 2002; Rolland et al., 2006). By using two copies of the CaMV 35S promoter in tandem to drive expression of the Grifola frondosa trehalose synthase gene in S. officinarum, Zhang et al. 2006 achieved trehalose accumulation in sugarcane cells at 0.9–1.2% of fresh weight. This low-level production in transgenic plants has been seen in other plant species and has been attributed to native trehalase activities (Garg et al., 2002; Penna, 2003). The transgenic lines showed no negative effects on growth, were drought tolerant, and produced higher yields under drought conditions.
2.2.7 Flowering
In sugarcane cultivars, the time and intensity of flowering are important as they influence the yield and quality of cane (Premachandran, 2006). Sugarcane yield is reduced by cessation of vegetative growth in flowering canes and profusely flowering clones are undesirable for commercial cultivation, especially when used for late-season crushing.
The identification of genes with major effects on flower induction could lead to new ways to control flowering in sugarcane cultivars (Ulian, 2006). Flower formation depends on the transition of a vegetative meristem to an inflorescence meristem and, subsequently, to a floral meristem. This transition requires the action of a number of genes, including LEAFY (LFY), a gene that is expressed early and considered to be important in the flowering process (Weigel et al., 1992). LFY interacts with another floral control gene, APETALA1 (AP1), to promote the transition from inflorescence to floral meristem. The ectopic expression of LFY induces the ectopic expression of AP1 in leaf and axillary flower primordia (Parcy et al., 1998). LFY and AP1 are pivotal for the switch to the reproductive phase, where, instead of leaves, the shoot apical meristem produces flowers. The AP1 promoter is a direct target of LFY (Wagner et al., 1999). LFY also directly targets the AGAMOUS (AG) promoter that contains a LFY-responsive enhancer necessary for its activity (Busch et al., 1999). AG encodes a transcription factor that regulates genes determining stamen and carpel development in wild-type flowers (Yanofsky et al., 1990). LFY, therefore, plays pivotal roles in the specification of flowers and in the patterning of floral organs (Hempel et al., 2000).
To study flowering in sugarcane, Ulian 2006 analyzed the SUCEST database and found a DNA EST with significant similarity to the Arabidopsis LFY sequence with two regions highly conserved in the LFY family of transcription factors. The putative LFY gene from sugarcane was expressed in the antisense orientation in transgenic sugarcane clone SP87-432 under the control of the constitutive maize Ubi-1 promoter. Ulian 2006 observed that silencing of the LFY gene suppressed flowering in transgenic sugarcane, supporting the notion that the gene plays an important role in sugarcane flower development.
3 Future Road Map
Recent research on sugarcane has started to produce the detailed biochemical and genetic information that will be needed to develop technologies necessary for sugarcane improvement and to metabolically engineer sugarcane for various traits and compounds.
3.1 Expected Technologies
To successfully manipulate the metabolic processes in sugarcane, a combination of molecular tools is required. In addition, consideration has to be given to both the organizational complexity at the whole plant level and the metabolic compartmentation within cells. Metabolic engineering requires a transformation system, suitable gene(s) and vector constructs, appropriate promoter sequences for cell- and tissue-specific expression, and effective targeting signals to direct the protein to its final destination within the cell. In sugarcane, an efficient transformation system (Bower and Birch, 1992; Bower et al., 1996) and effective targeting signals to direct heterologous proteins to various compartments including the vacuoles, plastids, and mitochondria are available (Gnanasambandam and Birch, 2004; McQualter et al., 2005; Rae et al., 2006; Petrasovits et al., 2007; Gnanasambandam et al., 2007). Future research should focus on improving the efficiency of existing technologies as well as developing new technologies.
3.1.1 Improved nuclear transformation efficiency
Currently, the biolistic method is the most efficient method for the production of transgenic sugarcane plants (Bower and Birch, 1992). However, transformed plants obtained through this method have several shortcomings, including the presence of numerous copies of transgenes with complex patterns of integration (Albert et al., 2003; Wang et al., 2003). This creates some difficulties in obtaining regulatory approval for transgenic crop plants. Multiple transgene integrations may also inhibit transgene expression, cause gene silencing, and/or promote transgene rearrangements. Additionally, biolistic transformation entails incorporation of the entire plasmid into the plant nuclear genome, which is a cause for public concern. Thus, genes essential for plasmid replication (ori) in bacteria as well as antibiotic resistance (e.g., ampicillin) required for selection of the transformed bacterial colony are co-introduced along with the gene(s) of interest. To minimize these problems, two technologies are currently being tested—Agrobacterium-mediated transformation (see Section 2) and the use of linearized plasmid DNA (LDNA) containing only the promoter-ORF-terminator cassette.
Agrobacterium-mediated transformation generally produces transgenic plants with fewer copies of the transgenes. Agrobacterium-mediated transformation of sugarcane has been reported in a few laboratories (Section 2). Most reports in sugarcane have shown the number of transgene copies to be generally between one and three (see Section 2). This may lead to higher and more stable expression of the transgenes as has been reported in rice (Dai et al., 2001) and maize (Shou et al., 2004) when compared to biolistic transformants. However, the stability of transgene expression using Agrobacterium, compared to biolistic transformation, has yet to be verified in sugarcane. Also, the efficiency of transformation is comparatively lower than that for microprojectile bombardment. Future research must aim to increase the efficiency of this transformation method in sugarcane.
In Agrobacterium-mediated transformation, transfer of genes of interest (present in the T-region of the Ti plasmid of Agrobacterium) into the plant nucleus usually occurs between the right and left T-DNA (transfer DNA) borders of the plasmid (Hellens et al., 2000). Generally, this region only contains the regulatory sequences along with the genes of interest. Thus, the transfer is believed to be precise, with none of the plasmid sequences flanking the border regions being transferred. Recently, however, Shou et al. 2004 analyzed transgenic maize to determine whether vector backbone sequences of the Ti plasmid were also transferred. They showed that in 75% of the R1 progeny of primary transformants there was some portion of the backbone present. This event may have occurred due to the specific transformation conditions used in the experiment. This needs to be determined when transforming sugarcane with Agrobacterium. Field trials of sugarcane plants transformed with Agrobacterium are proposed to commence in Australia in 2007 to address many of the concerns raised above (P. Joyce, personal communication).
Concerns raised by activists regarding the release of microbial genes into the environment coincidently with the biolistic transformation procedure has led scientists to use LDNA containing only the promoter-ORF-terminator cassette region for plant transformation. This LDNA method has been tested for gene expression and copy number analysis in rice (Fu et al., 2000; Loc et al., 2002). There was a significant reduction in the number of copies integrated into the rice genome. In addition, a low frequency of transgene rearrangements without any deleterious effect on expression pattern was observed (Fu et al., 2000). Moreover, when two minimal linear genes were co-bombarded, the co-transformation efficiency was similar to that using intact circularized plasmids. Similar experiments in sugarcane, however, showed that it behaved differently to rice (Albert et al., 2004; Joyce et al., 2006b).
3.1.2 Plastid transformation
The current nuclear transformation technology has several limitations, including semi-random and unpredictable incorporation of foreign genes into the genome, and the operation of genetic mechanisms that can switch off foreign genes, resulting in widely varying expression of introduced genes in sugarcane (Ingelbrecht et al., 1999; Gilbert et al., 2005; Vickers et al., 2005a, 2005b). A powerful new strategy, plastid transformation, has been developed in some plants, including Arabidopsis, tomato, potato, cotton, carrot, and rice (Bock and Khan, 2004; Koya et al., 2005), with the potential to circumvent these problems by introducing genes into plastid DNA rather than the plant nuclear genome (Maliga, 2002; Daniell et al., 2005b). The plastid transformation strategy utilizes two targeting sequences that flank the foreign genes and that inserts them through homologous recombination at a precise, predetermined location in the chloroplast genome (Maliga, 2004).
-
High levels of protein production: Because each leaf cell may contain up to 10000 copies of plastid DNA (each green cell contains about 50–100 chloroplasts with 60–100 plastid genomes per plastid), plastid-transformed tobacco plants synthesize extraordinary levels of foreign proteins (e.g., 5% of Bt toxin that provides resistance against insect pests (McBride et al., 1995)). This concentration is about 10-fold higher than the maximum achieved following transformation of the plant nuclear genome. These high levels of protein production in the plastids can facilitate the development of genetically improved sugarcane plants resistant to insect pests or herbicide damage, and the use of sugarcane plants as efficient bioreactors for production of novel high value biochemicals, pharmaceuticals, and industrial materials (Guda et al., 2000; DeGray et al., 2001; Daniell et al., 2005a).
-
Consistent gene expression: Protein expression from a gene inserted at a specific site in the plastid genome will be more consistent within the leaves of different transformants (Maliga, 2004).
-
Significantly reduced risk of spread of foreign genes (biocontainment): Chloroplasts are predominantly inherited through the maternal line in most important agricultural plants. Hence, the risk of a foreign gene being transmitted in pollen to nontransgenic sugarcane crops or related species is minimal (Lee et al., 2006). This has been experimentally shown by the containment of herbicide resistance in tobacco (Daniell et al., 1998)
-
Absence of gene silencing: Foreign genes can be silenced in sugarcane plants by modification of the inserted DNA in the nucleus, or by specific breakdown of the corresponding mRNA required as a template for protein synthesis in the cytosol. As there are no known similar silencing mechanisms identified in plastids (Dhingra et al., 2004; Lee et al., 2003a), foreign genes should be exempt from silencing in plastid transformed sugarcane plants, in which both transcription and translation occur within the chloroplast, separated from the cytosol.
-
The ability to express polycistronic messages from a single promoter: In nuclear transformation, each encoding sequence must be engineered under the control of a separate regulatory region, i.e., a monocistron. As a consequence, gene expression levels vary widely among introduced sequences, and generation of a number of transgenic plant lines is required to introduce all of the cistrons into one plant and to get proper coordinated expression in the target biochemical pathway. In contrast, the functioning of the plastid genome (being prokaryotic in nature) permits simultaneous expression of two or more genes from a single plastid promoter region (Quesada-Vargas et al., 2005). Such an expression method makes possible large scale and inexpensive production of some proteins and fine chemicals. In addition, it allows the engineering of metabolic pathways through the introduction of multiples genes in a single operon.
-
Integration via a homologous recombination process that facilitates targeted gene replacement and precise transgene control.
-
Sequestering of foreign proteins in the organelle, which prevents adverse interactions with the cytoplasmic environment. In addition, the ability of chloroplasts to form disulfide bonds and to fold human proteins could allow high-level production of biopharmaceuticals in plants.
Despite its tremendous biotechnological potential, plastid transformation has only been used routinely in tobacco (Maliga, 2004) and attempts to develop plastid transformation in sugarcane have been unsuccessful to date. However, a recent patent issued on plastid transformation in monocots (Daniell, 2006) indicates the potential exists to develop the technology in sugarcane. The complete nucleotide sequence of the chloroplast genome of sugarcane has been determined recently (Asano et al., 2004). The plastome of sugarcane is a circular double-stranded DNA molecule, 141182bp in size, and is composed of a large single copy of 83048bp, a small single copy of 12544bp, and a pair of inverted repeat regions of 22795bp each. The sugarcane chloroplast genome is similar to maize, but not to rice or wheat. The availability of sequences for sugarcane chloroplast genome (Asano et al., 2004) could facilitate the development of plastid transformation technology in sugarcane.
3.1.3 Avoiding antibiotic resistance gene in transgenic plants
-
Use of no selectable marker gene, i.e., only the gene of interest is introduced and transformed tissue is screened for (not a practical solution due to time constraints).
-
Use of a visual marker gene such as GFP (which has no harmful biological activities) to select transformed cells (Elliott et al., 1998), which can be cumbersome and prone to contamination.
-
Selecting T1 progeny that do not contain the antibiotic resistance gene, i.e., they have segregated for the antibiotic resistance gene. This is not an option for sugarcane, because none of the commercial sugarcane cultivars are homozygous. Sugarcane clones are clonally propagated to maintain the elite characters.
-
Use of an antibiotic gene excision system; once again, this will involve retransformation of transgenic cane. However, if more inducible promoters were available, it would be possible to control excision of the antibiotic gene (after selection and regeneration of plants) by switching on the expression of the site-specific recombinase gene (Puchta, 2000).
-
Use of a selection system that is not antibiotic based. The use of alternative marker genes conferring positive selection. For instance, a nontoxic compound to promote the regeneration and growth of transformed cells expressing a transgene that acts upon that compound are being developed. It may even be possible to tailor-make genes that are designed to specific plants.
-
Sugarcane plastid transformation would be one possible way of removing both antibiotic resistance genes and vector sequences from plants using homologous recombination.
-
Bombardment with linear DNA avoids vector-localized genes such as the ampicillin resistance (see Section 3.1.1). Transformation of sugarcane with linearized DNA fragments has been demonstrated (Joyce et al., 2006b).
3.1.4 Suppressing transgene silencing
There are convincing reports indicating that sugarcane displays a high level of PTGS (Liu et al., 2003; Wei et al., 2003). Strategies to suppress this include the use of viral suppressor proteins (Mangwende et al., 2005), addition of putative matrix attachment regions (MARS) (Wei et al., 2003), and presence of introns. The use of viral suppressors, including P0 from ScYLV and HcPro from SrMV, to advance our knowledge of RNA silencing in sugarcane are being explored (Wang et al., 2006, 2007). More research is needed to understand and suppress transgene silencing.
3.1.5 Tissue-specific promoters
Despite several years of research, an effective stem-specific promoter is not yet available in sugarcane. Though a stem-specific promoter has been reported in sugarcane (Hansom et al., 1999), its usefulness remains to be established. Similarly, other tissue-specific promoters that are operative in sugarcane (for example, root-specific promoters) are not available. Attempts to find new promoters for strong expression especially in the mature parenchyma cells of sugarcane stem are ongoing. Recently, Mudge et al. 2006 reported the isolation of an EST clone (SMS04) that was selected based on its expression pattern. Further analysis revealed that at least eight variants of this gene existed in sugarcane and that no two had the same sequence. The promoters associated with these genes were isolated and activity tested using the GUS reporter gene. Most were functional soon after introduction into the explant, but none maintained activity in mature regenerated plants.
To avoid gene silencing, Potier et al. 2006 isolated tissue-specific promoters from the upper stem or roots of maize and sorghum rather than sugarcane. These promoters (with and without the first intron) were used to test the expression of GUS gene in sugarcane callus. In general, the presence of an intron showed higher expression in all the promoters studied. Further research should identify effective tissue-specific promoters that can maintain activity in mature transgenic plants. With the available sorghum genome sequences, the search and solution to finding new tissue-specific promoters is more tangible.
3.1.6 Transcriptional regulators
Transcription factors are proteins that interact with the promoter regions of target genes in a sequence-specific manner. They influence the manner in which RNA Polymerase II initiates mRNA synthesis by enhancing or repressing gene expression and, hence, are important regulators of gene expression. Transcription factors tend to control multiple steps in metabolic pathways, meaning that a single protein can affect the expression pattern of a large number of downstream genes (Kinney, 2006). This is an important consideration for plant transformation strategies aimed at significantly affecting the level of end-product accumulation. Although this can be a successful strategy, a lack of knowledge about a particular pathway may mean that additional and hitherto unknown rate-limiting steps may be present that may frustrate the attempt to significantly modify end-product accumulation. Overexpression of transcription factors holds the promise of significantly altering the overall flux through a metabolic pathway by controlling the transcription of multiple genes. The potential of using transcription factors to enhance current and future work into the genetic enhancement of sugarcane is highlighted in Sections 2.2.6, 3.2.6, 3.2.7, and 3.2.9.
3.1.7 Modifying more complex traits
Very few traits that are simply inherited have been described in sugarcane (Hogarth, 1987; Daugrois et al., 1996; Mudge et al., 1996a, b; Raboin et al., 2006), suggesting that most phenotypic characteristics, such as cane yield and sugar content, are controlled by polygenes. In other organisms, genes that contribute to complex traits (QTL) pose special challenges that make gene discovery and subsequent modification of such traits more difficult. Such problems include locus heterogeneity, epistasis, low penetrance, variable expressivity, and pleiotropy (Glazier et al., 2002). Approaches are even more difficult in sugarcane, where there is the possibility of segregation for three or more alleles at a locus, a lack of chromosome preferential pairing and high levels of heterozygosity (Ming et al., 2001a, b; Aitken et al., 2006), which generally results in numerous alleles of small effect, some of which cannot be positioned on a genetic map. These factors work together to make claims of linkage discovery notoriously difficult to verify and candidate gene identification challenging. However, prospects for success can be improved through genome sequencing and comparative genomics. Such resources will allow more rapid dissection of complex traits through candidate gene identification and subsequent testing using functional genomics approaches.
3.1.8 Functional and comparative genomics
3.1.8.1 Expressed sequence tags (ESTs)
Sequencing efforts by different organizations in several countries have provided a large set of publicly available ESTs in sugarcane (Carson and Botha, 2000; Casu et al., 2003; Vettore et al., 2003; Ma et al., 2004). The largest of these efforts was undertaken in Brazil by the Organization for Nucleotide Sequencing and Analysis (ONSA). A network of laboratories in all the major research institutions across the state of São Paulo conducted genomics and transcriptomics (Harvey and McMeekin, 2005) on sugarcane ESTs (SUCEST) (http://sucest.lad.dcc.unicamp.br/en). The goal of the SUCEST project was to identify 50000 sugarcane genes or generate a total of 300000 ESTs. Following sequencing, the data mining groups in SUCEST worked on a cross-section of sugarcane genes including, but not limited to genes involved in amino acid metabolism, carbohydrate metabolism, lipid metabolism, energy metabolism, metabolism of co-factors (vitamins and other substances), nutrient uptake and metabolism, environmental stress, pathogenesis, membrane receptors, phytohormone biosynthesis and regulation, plant morphology, flowering and development of reproductive organs, patterns and levels of gene variation, and comparative genomics.
The outcomes of the SUCEST project were significant. It established Brazil as a biotechnology innovator, developed skills and capabilities throughout research institutions in São Paulo, created international collaborations and networks, and resulted in the formation of new biotech companies (Harvey and McMeekin, 2005). Data from the SUCEST project, and from other sequencing efforts, been centralized at the Institute for Genomic Research's (TIGR) Saccharum officinarum Gene Index (SoGI) (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=s_officinarum; Quackenbush et al., 2001). TIGR aims to integrate research data from international sugarcane EST sequencing and gene research projects. Its ultimate goal is to represent a nonredundant view of all sugarcane genes and data on expression patterns, cellular roles, functions, and evolutionary relationships. The TIGR database has 255635 ESTs with a total of 78547 unique sequences. This database was recently moved to the Computational Biology and Functional Genomics group at Harvard University, where it is incorporated into an even bigger gene index project: http://compbio.dfci.harvard.edu/tgi/plant.html.
A subset of the available EST sequences have been arrayed on a Sugar Cane GeneChip® (Affymetrix) (Casu et al., 2006), which is designed specifically to monitor gene expression. The GeneChip® Sugar Cane Genome Array contains 8236 S. officinarum probe sets to monitor gene expression for approximately 6024 distinct genes. Expression profiling using this and other cDNA arrays, along with mapping studies, have investigated genes involved in stem development, sugar accumulation, growth at low temperatures, and disease resistance (Casu et al., 2003, 2004, 2005, 2006; Nogueira et al., 2003; Rossi et al., 2003). These, and future studies, should provide targets for functional genomic screens through either overexpression or RNAi-mediated gene knockout strategies. In fact, field testing of a number of lines for increased sugar yield has already begun by the Centro de Tecnologia Canavieira (CTC) and Allelyx, a biotech start up company developed out of the ONSA genomics program (Harvey and McMeekin, 2005).
Importantly, the wealth of sequence information available allows for a robust strategy to incorporate all polymorphic alleles. The EST database is also a starting point for metabolic engineering by helping to understand sugarcane central metabolism. For instance, in a recent approach to produce trehalose in sugarcane, a survey of the EST collection identified a probable trehalose dehydrogenase, the activity of which would have to be inhibited for the strategy to be successful (O'Neill et al., 2006).
3.1.8.2 Genomics
The large database of public ESTs described above is an important resource that has been useful in a variety of ways, providing functional DNA markers, a foundation for development of expression profiling platforms, and a rich resource for evolutionary studies. Available EST sequences may collectively provide tags for half of the sugarcane genes. However, genomic DNA-based systems are also required because (i) many important genes expressed at low levels are unlikely to be found in libraries and (ii) the cDNA approach provides no information about the crucial on/off switches (promoters, enhancers, transcriptional regulators) that control gene expression. Hence, a comprehensive picture of the sugarcane genome including the entire suite of genes, their all important regulatory elements, and their complete arrangement along the chromosomes will eventually require complete sequencing of the genomes of one or more sugarcane genotypes. Ultimately, this could be achieved with whole-genome sequencing.
A major step toward full genome sequencing would be the construction of large sugarcane bacterial artificial chromosome (BAC) libraries and the generation of detailed genomic maps. Brumbley and Brumbley 2005 generated a library of sugarcane cultivar Q200A containing 98000 colonies with an approximate 1 × coverage of the genome. Larger libraries of sugarcane cultivar R570 were constructed at Clemson University and at CIRAD in Montpelier, France. Rapid progress in the speed and cost of DNA sequencing will make feasible the sequencing of entire sugarcane genomes within a few years (Paterson, 2006). Once sequenced, the door is open for candidate gene studies using functional genomics. In the interim, much is likely to be learned from comparative genomics studies using the sequences of closely related sorghum genome both as a source of control elements and/or as a tool to identify likely candidate genes through comparison of QTLs on sugarcane genetic maps with the physical map of sorghum (Guimarães et al., 1997; Ming et al., 1998).
Sorghum is representative of tropical grasses in that it is classified as a C4 photosynthesizer, comprising complex biochemical and morphological specializations that improve carbon assimilation at high temperatures. By contrast, rice is more representative of C3 temperate grasses. Sorghum's economic and scientific importance, together with progress in characterizing its genome (detailed below) have motivated the sequencing of an elite inbred line of S. bicolor L. genotype, BTx623, to 8x genome coverage under the US Department of Energy Joint Genome Institute (JGI) “Community Sequencing Program” (CSP). Sorghum sequencing was completed in early 2007 and the database is available at the same Harvard University site as sugarcane, http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=sorghum, and annotation is well underway.
The completed sorghum sequence will be an excellent bridge for translating hard-won functional genomics knowledge from Oryza sativa L. ssp. indica to leading tropical grass crops, with much larger genomes and much more gene duplication. Sorghum and maize diverged from a common ancestor approximately 12 million years ago (Gaut et al., 1997; Swigonova et al., 2004a), versus approximately 42 million years for rice and the maize/sorghum lineage (Paterson et al., 2004). Saccharum may have shared ancestry with sorghum as little as 5 million years ago (Sobral et al., 1994) and it retains a similar gene order (Ming et al., 1998), and even produces viable progeny in some intergeneric crosses (Dewet et al., 1976). Maize has undergone a whole-genome duplication since its divergence from sorghum (Swigonova et al., 2004b), and Saccharum has undergone at least two (Ming et al., 1998). The low level of gene duplication in sorghum makes it, like rice, an attractive model for functional genomics.
The sorghum sequence will also offer numerous direct opportunities to increase knowledge of sugarcane biology. At the level of genome structure, sorghum and sugarcane chromosomes appear to share very similar gene content and order, and it is expected to be simple to use well-established synteny relationships (Ming et al., 1998), supplemented by a growing body of hybridization marker and end-sequence data to deduce the likely locations in sugarcane of genes identified in the sorghum sequence. Its much closer relationship will make the sorghum sequence of substantially greater value than the rice sequence for such deductions in sugarcane.
A host of classical genetics, supplemented by recent DNA marker studies (Ming et al., 2001a, 2001b, 2002a, 2002b) show heterozygosity resulting from autopolyploidy to be instrumental in the productivity of sugarcane. More generally, genome duplication has long been suggested to play an important role in angiosperm evolution (Stephens, 1951; Ohno, 1970). One of the biggest surprises of the genomics era has been underestimation of the prevalence of ancient whole-genome duplication, finding that even the genomes of diploids such as Arabidopsis (Blanc et al., 2000; Initiative, the Arabidopsis Genome, 2000; Paterson et al., 2000; Bowers et al., 2003) and Oryza (Paterson et al., 2003, 2004, 2005; Vandepoele et al., 2003; Wang et al., 2005b) have been shaped by genome duplication. Such duplication is associated with a host of rapid responses including loss and restructuring of low-copy DNA sequences (Song et al., 1995; Feldman et al., 1997; Ozkan et al., 2001; Shaked et al., 2001; Kashkush et al., 2002), activation of genes and retrotransposons (O'Neill et al., 1998; Kashkush et al., 2003), gene silencing (Chen and Pikaard, 1997a, b; Comai, 2000; Comai et al., 2000; Lee and Chen, 2001), and organ-specific subfunctionalization of gene expression patterns (Adams et al., 2003, 2004). These mechanisms may be especially important in providing raw material for evolutionary change in sugarcane, in that it appears to have undergone two such genome duplications in about the past 5 million years or so. Sorghum will provide an ideal outgroup for deducing the ancestral states of sugarcane genes that have been affected by genome duplication, in that it is very closely related but diverged prior to the duplication events. Future comparison of the fates of genes in sugarcane to those in Sorghum halepense, a polyploid derivative of sorghum, may offer the opportunity to test hypotheses about the degree to which the fates of duplicated genes are determined by a taxon-independent set of molecular principles (Paterson et al., 2006). This could have profound implications for transgenic strategies in the future. For a given trait, are single copies or multiple copies of transgenes more beneficial in sugarcane?
3.2 Expected Products
The future for sugarcane biotechnology is exciting. Because of its narrow genetic base, there is enormous potential to improve the field performance of this crop. Sugarcane is considered to be an ideal biofactory for the production of a range of sugars, and fine chemicals, including biopolymers, nutraceuticals, industrial enzymes, and pharmaceuticals, for the following reasons: (i) sugarcane is one of the fastest growing and largest biomass-producing agricultural crops in the world; (ii) it accumulates sucrose up to 42% of the dry weight of the stalk. In addition, bagasse (fibrous residue of sugarcane stalks left after extracting the sucrose) is made up of 32–48% cellulose, 19–24% hemicellulose, and 23–32% lignin, all of which all have commercial uses; (iii) it is grown throughout the tropical and subtropical latitudes and is a major crop in many developing nations and can, therefore, be used to help developing economies; (iv) it is usually vegetatively propagated and only produces fertile seed under a specific set of environmental conditions, therefore posing a low risk for transgene flow into the environment; (v) sugarcane crop-cycles run for 4 or more years between plantings. Hence, it does not have to be replanted every year, as it produces new tillers from the stool left in the ground after harvesting; (vi) sugar industries throughout the world already have substantial infrastructure to harvest the large biomass and transport it to mills for processing; and (vii) the tops and leaves of sugarcane, which are traditionally burnt or left behind in the field as a trash blanket, could potentially be used as a production platform for high value biofactory products, as another source of biomass for biorefineries or, more likely, both. In the future, it is highly likely that transgenic sugarcane will be grown not only for the production of sugars for human consumption, but also as a high biomass crop for fuel and energy production, with value adding by metabolically engineering it to produce a range of additional products in planta.
3.2.1 Improved sugar
Sugarcane is unusual amongst plants in storing up to 62% dry weight (and 16–25% fresh weight) of sucrose as the primary source of carbon and energy reserve in the storage parenchyma of the mature culm or stalk (Bull and Glasziou, 1963). These are approximate levels obtained in S. officinarum, the major source of commercial hybrid germplasm. In contrast, some of the wild relatives of sugarcane store less than 2% of the fresh weight as sucrose. As the photosynthetic rates on an area basis of S. spontaneum have been reported as nearly twice those of S. officinarum and 30% greater than that of hybrid cultivars (Irvine, 1975), these striking differences in sink accumulation and storage activity cannot be explained by differences in photosynthetic rates in the leaves (Moore, 2005). Despite many years of research, the process of sugar accumulation is not fully understood.
As more than 70% of the sugar harvested for human consumption is derived from sugarcane (FAO, 2006), improving sugar content in sugarcane will remain as an important focus for future biotechnological innovations. Traditional breeding has improved the yield of sucrose mostly through increased cane yield. Increasing sucrose concentration has not occurred recently through traditional breeding in countries such as Australia, where the levels are already relatively high (Jackson, 2005). Attempts to increase sucrose content in sugarcane cultivars through the modification of plant genes involved in sugar metabolism have been mostly unsuccessful (reviewed by Lakshmanan et al., 2005). For example, a 70% reduction in activity of acid invertase, a key enzyme involved in sucrose breakdown, resulted in no significant change in sucrose yield or purity in the immature internodes of transgenic sugarcane (Botha et al., 2001). A better understanding of sugar-transport pathways will be required for strategies to increase the flux of sugars and, ultimately the amount of sucrose in the storage tissue (Grof and Campbell, 2001; Rae et al., 2005).
The doubled sugar content observed in transgenic sugarcane while attempting to produce isomaltulose (Wu and Birch, 2007) indicates that there is potential to improve sugar content in sugarcane through metabolic engineering of the vacuolar compartment. However, most vacuolar-targeting studies in many crops involving stable transformants have been limited by difficulties in detecting vacuolar-targeted enzyme activities (Ebskamp et al., 1994; van der Meer et al., 1994; Caimi et al., 1996; Ma et al., 2000; Gnanasambandam and Birch, 2004), indicating that the stability of targeted proteins is an important factor for effective metabolic engineering. For isomaltulose production in sugarcane, a highly efficient sucrose isomerase had to be used (Wu and Birch, 2007).
Reports of experimental efforts to protect foreign gene products from vacuolar degradation are scarce. To understand how to genetically engineer a protein for stability in the vacuole, the “enemies” within the vacuoles—the proteolytic enzymes and their sites of action—need to be characterized (http://www.proweb.org/other.html). A method to isolate protoplasts from sugarcane mature stem parenchyma cells has been reported (Gnanasambandam and Birch, 2006), but further optimization is needed for successful vacuole isolation for characterization of proteolytic enzymes. Alternatively, if vacuole isolation proves to be difficult, fluorescent substrates for proteases (from Molecular Probes, Inc.) can be used with isolated protoplasts. Such characterization on proteolytic enzymes may allow for informed modification on foreign proteins to remove sites for attack by sugarcane proteases. Alternatively, directed evolution can be applied to rapidly evolve enzymes for optimal activity under vacuolar conditions (Stemmer, 1994a, 1994b; Chica et al., 2005; Kaur and Sharma, 2006). There may be particular sequences within a protein that are sensitive to proteolytic enzymes, and altering these sensitive sequences may increase protein stability. For example, mutation of protease-sensitive regions of the firefly luc yielded larger and more stable signals compared with the wild-type protein in E. coli (Thompson et al., 1997).
3.2.2 Alternative or novel sugars
There have already been examples of alternative sugars produced in genetically modified (GM) sugarcane (Section 2). In addition to isomaltulose (Wu and Birch, 2007) and sorbitol (Chong et al., 2007), there is potential to engineer synthesis of a range of other sugar complexes and sugars into sugarcane, for example alternative carbohydrates such as starch and fructans (already demonstrated in sugarbeet).
Tagatose (Kim, 2004) has properties closer to sucrose than any of the other sugar substitutes. D-Tagatose is an isomer of D-galactose and it can function as a low-Joule sweetener. Although it has relatively the same sweetness as sucrose, it is detected on the tongue sooner.
Xylitol is a sugar alcohol of considerable commercial interest (Granström et al., 2007a, 2007b). Its metabolism in humans is independent of insulin and it is anticariogenic. It also has uses as a pharmaceutical. Xylose is a major component of sugarcane hemicellulose, making up 25% of the w/w dry matter and 91% of the nonfermentable sugars (Pessoa Jr. et al., 1997). Research is currently underway to optimize the conversion xylose to xylitol. Candida guilliermondii has been used as a test species on sugarcane bagasse hydrolysate, because it naturally metabolizes xylose (da Silva and Felipe, 2006). However, Candida cannot be used in food production, because many Candida species are pathogens. An alternative is to use bacteria. When a xylose reductase and a xylose transporter were engineered into the bacterium Lactococcus lactis, levels of xylitol production were equivalent to those in the best-producing Candida line (Nyyssölö et al., 2005). An alternative would be to engineer a xylose reductase into sugarcane. One strategy might be to have the enzyme incorporated into the sugarcane cell wall structure, so that it is only functional when it is released during sugarcane processing at the mill. Another might be to regulate it with a wound-inducible promoter, so that it is produced in the sugarcane cells after harvesting. In both of these scenarios, the enzyme(s) will have to be engineered to function at the temperatures and pH of a sugar mill and in particular under mill conditions designed to break down hemicellulose (Pessoa Jr. et al., 1997).
Though there has been much research toward the development of low-Joule nonsucrose sweeteners (Modi and Borges, 2005), two such sweeteners (saccharin and cyclamates) were found to be carcinogenic under some circumstances, while others (such as aspartame) exhibit temperature and low pH instability that limit their utility (Knight, 1994). Hence, it may be useful to produce a low-Joule nonsucrose product from sugarcane. Alternatively, there are a range of sweet proteins that may be worth considering. Monellin, Thaumatin, and Brazzein are 100000, 3000, and 500 times sweeter than sucrose, respectively. These would be a natural replacement for synthetic sugars because they are low-Joule sweeteners and would not trigger insulin production (Kant, 2005).
3.2.3 The biorefinery—biofuels and biochemicals
Sugarcane is expected to become a platform for biorefinery production of fuels and chemicals. Recent demand for renewable energy sources has increased the interest within the research community on transgenic breeding for biomass production. Sugarcane is the crop having the most favorable input/output balance for bioenergy, and with the growing body of research on its genome, may allow the development of a low-input energy crop with key outputs as biofuel (ethanol) and bioenergy (electricity). The production of an alternative energy crop, with high biomass potential, will be a significant step toward the implementation of a biofuels, bioenergy, and, in the not too distant future, bioproducts industries. If biorefineries, producing dozens of products (Figure 6) are integrated into sugar mills, one can envision a sugarcane industry functioning more like petrochemical plants do today. Unlike petrochemical plants, the resource driving this new bioeconomic revolution will be renewable.
In nature, the most prevalent forms of carbon are the biopolymers cellulose and hemicellulose (Houghton et al., 2006). The third most abundant biopolymer is lignin (van Dam et al., 2005). These are considered to be the raw materials for future bio-based industries creating opportunities for sugarcane industries to diversify. Aside from water, sugarcane plants are primarily comprised of cellulose, hemicellulose, and lignin. Traditionally, this resource has been combusted, either in the field to remove the leaf material or at the mill to utilize the waste product bagasse to generate the necessary energy to power mill operations. Since the mid-1980s, it has been proposed that “energy cane” should be introduced as a multiple product alternative to sugarcane grown largely for sucrose (Alexander, 1984, 1985). The time may have come to implement this vision with the inclusion of advances in the fields of biotechnology. Changing the sugarcane industry from primarily a one-product to a multiple-product industry is an enormous challenge, but there are multiple indicators that should make both sugarcane farmers and millers optimistic.
Of all the agricultural crops, sugarcane may be the most ideally suited to benefit from a shift to a bio-based economy because of its large biomass and, therefore, large quantities of cellulosic and hemicellulosic sugars and lignin (Alexander, 1984, 1985; Brumbley et al., 2004, 2006b, 2006c, 2007). There is good reason for simultaneously developing sugarcane both as a biofactory and as a feedstock for biorefinery processes. As technologies are developed to produce a range of new and diverse products in the sugarcane plant, other technologies are being developed to dismantle the cellulose and hemicellulose to release the large quantities of fermentable sugars from biomass. The breakdown of the tough structural elements that make up sugarcane cell walls will facilitate the extraction of new bioproducts such as the bioplastics described in Section 2.
The biofuel ethanol is becoming increasingly more important as the world attempts to wean itself off its addiction to petroleum-based fuels. The second largest producer of ethanol in the world is Brazil, and sugarcane is their feedstock (Pessoa Jr. et al., 2005; Lorentzen, 2006). In the near future, another bio-based fuel (butanol) may enter the market. DuPont and British Petroleum recently formed a joint venture to produce this biofuel commercially. Butanol resolves many of the problems of ethanol. Not only does it have a higher energy content, it is not as corrosive, uses an air/fuel ratio that is closer to that of gasoline, can be shipped through existing fuel pipelines, is safer to handle, and can replace gasoline at any percentage up to 100%.
Governments and private industry are investing in technologies to convert biomass into biofuels and biochemicals. Cellulose, hemicellulose and lignin interact to form plant cell walls and are responsible for giving plants their structural integrity. Compared to starch, getting to the sugars that make up these polymers is far more difficult and more expensive (Bayer et al., 2004; Houghton et al., 2006). A diverse range of bacteria have been discovered that have multienzyme complexes that bind to and degrade cellulose and hemicellulose. This complex has been termed the cellulosome and the research on these enzyme complexes was reviewed recently (Bayer et al., 2004). Researchers are looking for new organisms with cellulose degrading capabilities. Once discovered, activity and performance of these enzymes and complexes can be improved using directed evolution (Joyce, 2004; Neylon, 2004; Hibbert and Dalby, 2005).
Potential product targets for sugarcane-based biofactories and biorefineries. From just a few building blocks an array of secondary and intermediates chemicals can be produced. The intermediate chemicals are the building blocks of a vast range of end product.
[Reproduced with permission from Todd Werpy and Gene Petersen, the lead authors of “Top Value Added Chemicals from Biomass”]
The pool of sugars currently locked up in cellulosic biomass could be used to feed biorefineries both for the production of biofuels and bioproducts. White biotechnology (Paster et al., 2003) is the use of biological processes for the production of chemicals such as plastics, adhesives, and paints. Fermentation technologies could be used to produce building block chemicals that will be the basis for the range of secondary and intermediate chemicals (Figure 6). Currently, the United States and European Union annually produce €1000 billion worth of fine and bulk chemicals from a petroleum feedstock. They plan to shift over 90% of this production from a petroleum to a biorenewable base over the next 50 years. To accomplish this, new enzymes will be required to convert the sugars within the cellulosic biomass to the renewable building blocks required by tomorrow's manufacturing sector.
The vast majority of microbes have never been characterized. This enormous and diverse population is thought to be a rich source of new genes, metabolic pathways, and enzymes. However, most of these organisms are not culturable. Metagenomics is the study of the collective genomes of organisms, independent of growth in culture. The tools for doing metagenomics are advancing rapidly (Tringe and Rubin, 2005; Green and Keller, 2006). For example, Venter et al. 2004 used metagenomics to identify over 1.2 million new genes from at least 1800 species, including 148 previously unknown bacterial groups, just in the Sargasso Sea. As our understanding of protein domains advances (Portugaly et al., 2007), it may be possible to design “lab-on-a-chip” devices (Hong et al., 2004) that can be used to rapidly screen different environmental samples for genes encoding specific enzyme domains. Using systems biotechnology, newly discovered enzyme's can be characterized rapidly and their utilities for industrial bioprocessing determined (Lee et al., 2005). Directed evolution tools, such as domain shuffling (Hibbert and Dalby, 2005), can be utilized to combine the new enzyme's functional domains with those of other related enzymes to create an enzyme that will function in the precise environment it is needed, whether that is a fermentation tank inside the cytoplasm, storage vacuole, mitochondria, peroxisome, or plastid of a sugarcane cell.
3.2.4 Biopolymers
A potential major industrial use for sugarcane is the production of biodegradable polymers. These polymers can be produced by fermentation technologies and, as shown in Section 2, can be produced in sugarcane. For example, PHAs are polyesters synthesized naturally by many species of bacteria as carbon sources and energy reserves. PHAs are accumulated to levels as high as 90% of the cell dry weight in bacteria. PHAs have the properties of thermoplastics and elastomers and they are biodegradable (Madison and Huisman, 1999). These classes of biopolymers offer a renewable and environmentally friendly alternative to the 150t per year of plastic currently produced from petrochemical resources.
In nature, over 130 PHA variations have been identified that produce plastics with properties varying from hard and brittle, too elastomeric, gluelike, and even rubbery (Steinbüchel and Valentin, 1995). PHAs are classified based both on the length of the side chains on the polymer (i.e., whether the side chains are short (C4-6) or medium (C6-16) in length), and on whether the polymer is a homopolymer, or a co-polymer of PHAs with different side chains. In the case of co-polymers, the ratio of medium (mcl) to short side (scl) chains also affects the properties. Co-polymer PHAs of commercial interest range from 2% mcl (hard, brittle) to 20% mcl (soft, elastic). PHB, the most studied of the PHAs, is a short-side-chain homopolymer and displays properties similar to polypropylene (Hocking and Marchessault, 1994).
In Brazil, an important precedent has been established on how to diversify the product base of a sugar industry, first with ethanol, and then with bioplastics (Lorentzen, 2006; Velho and Velho, 2006). In 1992, initial projects were funded to develop a novel platform for the production of PHB from sugarcane through fermentation (Velho and Velho, 2006). The Brazilian company Copersucar (http://www.copersucar.com.br) first engineered the bacterium Ralstonia eutropha to use sucrose as a carbon source. A pilot plant was then built, integrated into a sugarcane mill (http://www.biocycle.com.br), and ultimately produced 50t per year of high purity PHB. This refinery receives its feedstock (sugar syrup) directly from the sugar mill, which also produces all of the energy necessary to run the fermentation, extraction, and purification facilities. The solvents for PHB extraction are also sugar mill products sourced from the ethanol production facility (Velho and Velho, 2006). This model is an excellent example of how an existing resource industry can, with government and corporate support, build on its knowledge base to create new business opportunities and a help establish bio-based economy in a developing country (Lorentzen, 2006).
BiopolTM (Choi and Lee, 1999) is the only PHA co-polymer that has been produced commercially. However, Metabolix, Inc. has formed a strategic alliance with Archer Daniels Midland (ADM) to commercialize PHA natural polymers. Metabolix and ADM claim that they can make these natural plastics by fermentation at costs that are competitive, or near competitive, with petrochemical-based plastics (http://www.metabolix.com). Metabolix argues that producing these plastics in the cells of living plants will reduce this cost even further. Hence, agricultural crops are regarded as a promising low-cost alternative for the production of PHAs on a large scale (Snell and Peoples, 2002). High-level production (approximately 40% of dry weight) of PHB has been achieved in plants by introducing three R. eutropha genes encoding a β-ketothiolase (phaA), an acetoacetyl-CoA reductase (phaB), and a PHA synthase (phaC) (Bohmert et al., 2000). The success in the production of PHB in sugarcane (Petrasovits et al., 2007; Purnell et al., 2007) indicates that other biopolymers can be produced in sugarcane.
3.2.5 Natural products/pharmaceuticals/proteins
There are opportunities to engineer the metabolism of sugarcane to increase production of natural products (Dixon, 2005). For instance, the most functionally and structurally diverse group of plant metabolites is the isoprenoids (also called terpenoids). These diverse compounds have a commercial value as flavors, pigments, polymers, or drugs (reviewed in Rodríguez-Concepción, 2006; Withers and Keasling, 2007). There are other good reasons to learn how to manipulate and/or regulate isoprenoid production in sugarcane, as they play primary roles in respiration, photosynthesis, and regulation of growth and development. As secondary metabolites, they function in protecting plants against herbivores and pathogens, attract pollinators and seed dispersing animals, and influence competition among plant species (Rodríguez-Concepción, 2006).
As described in Section 2, some work has already been done to engineer sugarcane to produce pharmaceuticals and high-value and bulk proteins. Monoclonal antibodies (mAbs) are one class of pharmaceutical proteins with considerable potential for production in plants. The estimated market size for mAbs is expected to exceed US $20 billion per year (Reichert et al., 2005). Both IgG- and IgA-type antibodies have been successfully produced in plants (Giritch et al., 2006). Recently a new transient expression technology, “magnifection”, for production of gram quantities (0.5gkg–1 fresh leaf biomass) of mAbs in plant cells was reported (Giritch et al., 2006). The time from gene delivery to production of fully assembled mAb is 14 days and to production of gram quantities is 14–20 days, making it the fastest production platform available (Hiatt and Pauly, 2006). This system uses vectors from two different noncompeting plant viruses, one for producing the heavy chain and one for the light chain, and transfers them to the plant via Agrobacterium-mediated transformation.
One area that may hold promise is the production of industrial enzymes. Future biorefineries are going to require a host of enzymes for converting base material into the raw materials required by the manufacturing sector (Figure 6). Some of the enzymes can be produced directly in the sugarcane cells, and potentially at high concentrations once plastid transformation technologies are developed for sugarcane.
3.2.6 Drought tolerance
In most crops, past breeding efforts for drought tolerance have been hindered by the quantitative genetic basis of the trait and the poor understanding of the physiology (reviewed in Tuberosa and Salvi, 2006). For breeders, it is important to select genotypes that are able to optimize water uptake and water use efficiencies and minimize damage incurred by drought stress. This ultimately will lead to maximized yields. In an effort to select for improved drought tolerance, breeders have identified a myriad of morpho-physiological QTLs acting from the cellular to the whole crop level (reviewed in Tuberosa and Salvi, 2006). However, given the genetic complexity of sugarcane, a marker-assisted selection approach for a myriad of improved drought tolerance QTLs would appear a very difficult proposition. An alternative way forward would be to make use of the knowledge gained in simpler systems and identify candidate genes to test in sugarcane. Candidate genes for drought tolerance have been divided into three broad areas including: (i) functional proteins such as enzymes involved in osmotically active compounds, transporters, chaperones, and reactive oxygen species (ROS) scavengers; (ii) transcription factors involved in the plant response to drought stress; and (iii) signaling factors upstream of transcription factors, including protein kinases and proteins involved in phospholipid metabolism, calcium sensing, and protein degradation (reviewed in Umezawa et al., 2006; Valliyodan and Nguyen, 2006). Most of these validation studies have been undertaken with model plants in laboratory or glasshouse conditions. An important next step is to trial these approaches in crop species under field conditions.
In other plants, a variety of transgenes have been used to improve drought tolerance. Examples include, the barley HVA1 gene (Fu et al., 2006), manganese superoxide dismutase (Wang et al., 2005c), regulator of G-protein signalling protein (RGS) (Chen et al., 2006), and aldehyde dehydrogenase (Rodrigues et al., 2006). Perhaps the most widely and successfully used genes have been the CBF/DREB group of transcription factors (Agarwal et al., 2006). CBF/DREB is a class of transcription factors that binds to drought responsive cis-acting elements and is important in regulating gene expression in response to drought, high salt, and cold stress (Yamaguchi-Shinozaki and Shinozaki, 1994). The ability of CBF to enhance tolerance to cold stress (Jaglo-Ottosen et al., 1998) and drought stress (Haake et al., 2002) was first demonstrated in Arabidopsis and has subsequently been shown to enhance tolerance to abiotic stress in commercially important crops such as tomato (Hsieh et al., 2002; Lee et al., 2003b) and rice (Ito et al., 2006).
Sugarcane lines have been produced containing a gene encoding drought-induced Arabidopsis transcription factor, AtCBF4 (McQualter and Dookun, 2007). Under glasshouse conditions, plants containing AtCBF4 driven by the constitutive maize polyubiquitin promoter showed growth retardation. Research is now focused on using drought-inducible promoters to reduce this negative phenotype.
As mentioned in Section 2, trehalose has already been used to engineer drought tolerance in sugarcane (Zhang et al., 2006). There are also a number of other potential osmoprotectants that can be trialed in sugarcane. These include betaines, the amino acids proline and ectoine, and polyols mannitol and sorbitol (Rontein et al., 2002). As mentioned above, transgenic sugarcane plants have been generated that produce sorbitol (Chong et al., 2007) and are being progressed to field trials.
3.2.7 Disease and insect resistance
Estimates of the cost of control and loss due to major sugarcane pests and diseases in Australia are in the order of 10% of the total value of the sugarcane crop (McLeod et al., 1999). Of these diseases and pests, soil-borne pathogens caused an estimated 75% of the losses, followed by canegrubs (larvae of Scarabaeidae) (10%), ratoon stunting disease (caused by Lxx.) (6%), and brown rust (caused by Puccinia melanocephala H&P Sydow) (3%). Occasionally, disease outbreaks due to the breakdown of disease resistance occur, which can result in much larger losses (40% reduction in cane yield in some sugarcane growing regions in Australia as a result of orange rust (Magarey et al., 2001)). Diseases such as ratoon stunting disease, pineapple disease, and red rot can be largely controlled by farm hygiene processes such as sterilizing cutting and harvesting equipment and by the use of fungicides (Magarey, 2005). However, for most sugarcane diseases, genetic resistance is the most effective method of disease management and sugarcane breeders have done an excellent job in controlling diseases through the use of resistant cultivars.
In the future, it is possible that resistance may break down, and aside from introgressing wild germplasm with novel resistance (if this exists in the gene pool), alternative strategies will be needed. Such approaches should provide durable resistance, preferably against a range of pathogens. Genetic engineering has the potential to achieve this through inserting carefully selected and possibly multiple genes as transgenes that (i) confer durable broad-spectrum resistance; (ii) are safe for all other organisms; and (iii) cause no yield penalty in the plant (Gurr and Rushton, 2005a). Such approaches have been successful with chewing insect pests, where expression of an insecticidal protein from B. thuringiensis has lead to increased yields and reductions in insecticide applications in cotton, soybean and maize (reviewed Gurr and Rushton, 2005b). Recent work in transgenic sugarcane has also indicated that Bt proteins are active against stem borers (Weng et al., 2006). Strategies for resistance to sugarcane viruses have also met with some success through RNAi-mediated approaches against the single-stranded RNA SCMV and the double-stranded RNA FDV (Joyce et al., 1997; Ingelbrecht et al., 1999; McQualter et al., 2004). However, similar successes for durable resistance to bacteria and fungi have not been forthcoming (reviewed Gurr and Rushton, 2005a).
Strategies have attempted to boost disease resistance through constitutive overexpression of defense components or single-antimicrobial proteins and have led to a variety of problems including poor quality plants, yield reduction, poor efficacy, or short durability (reviewed by Gurr and Rushton, 2005b). New strategies are being explored to overcome these problems. For example, a two-component system has been suggested, where the manipulation of “master switch” genes (such as kinases and transcription factors), which regulate entire signaling pathways are coupled with pathogen-inducible promoters to precisely regulate spatial and temporal gene expression (Yamamizo et al., 2006). Other strategies have come from the genomics field, for instance the complete genome of Lxx, has been fully sequenced (Monteiro-Vitorello et al., 2004) and opportunities now exist to try and control or even eradicate this costly pathogen from sugarcane (Brumbley et al., 2006a). Global transcriptional analysis of plant pathogen interactions has also revealed pivotal steps in defense responses that may be targeted in future strategies (Zabala et al., 2006). Further disease control strategies may involve pyramiding two or more antimicrobial proteins to improve efficacy and durability of resistance.
Work on transcription factors in other plants may offer some future promise for sugarcane. WRKY proteins are a large family of transcription factors that mainly participate in plant biotic-stress responses (Liu et al., 2007). They share a DNA-binding domain consisting of about 60 amino acids, as well as additional conserved features that distinguish individual subgroups (Eulgem et al., 2000). WRKY transcription factors can bind to domains in the promoters of pathogen-response genes, termed W boxes (TTGACC) (Maeo et al., 2001; Turck et al., 2004). While overexpression of some WRKY transcription factors has resulted in enhanced resistance to various bacterial or fungal pathogens, others can suppress the expression of defense-related genes.
Chen and Chen 2002 transformed Arabidopsis plants with AtWRKY18 under control of the CaMV 35S promoter. Transgenic AtWRKY18 plants showed marked increase in the expression of pathogenesis-related genes and resistance to the bacterial pathogen Pseudomonas syringae. Thus, AtWRKY18 was shown to positively modulate defense-related gene expression and disease resistance. Conversely, AtWRKY7 transcription factor plays a negative role in defense responses to P. syringae. Kim et al. 2006 studied the biological function of the Arabidopsis WRKY7 gene in both loss-of-function T-DNA insertion and RNAi mutants and gain-of-function transgenic overexpression plants. The T-DNA insertion and RNAi mutant plants displayed enhanced resistance to a virulent strain of the bacterial pathogen P. syringae. Transgenic Arabidopsis plants that constitutively overexpress WRKY7 showed reduced expression of defense-related genes, including PR1 and developed more severe disease symptoms than wild-type plants.
A large number of WRKY genes have also been identified in rice. Ryu et al. 2006 isolated WRKY transcription factors whose expressions were altered upon attack of the fungal pathogen Magnaporthe grisea, the causal agent of rice blast disease. Among 45 tested genes, the expression of 15 was increased in interactions between rice and M. grisea. Twelve of this subset of genes were also differentially regulated in rice plants infected with the bacterial pathogen X. oryzae pv. oryzae (X.o.o) 13751. Ryu et al. 2006 suggested that a large number of WRKY DNA-binding proteins are involved in the transcriptional activation of defense-related genes in response to rice pathogens. Several pathogenesis-related genes were induced in OsWRKY03-overexpressing transgenic rice plants (Liu et al., 2005), while overexpression of the OsWRKY71 gene in rice resulted in enhanced resistance to virulent bacterial pathogen X.o.o. 13751 (Liu et al., 2007).
As mentioned above, there are extensive databases at Harvard University covering plants, animals, fungi, and protists. The TIGR has the Comprehensive Microbial Resource (CMR) (http://cmr.tigr.org/). Within this free database are the complete genome sequences of 354 bacteria, including a number of plant pathogens. There is currently an effort to reduce the cost of sequencing the genome of a bacterium to under US $1000, and this is likely to be accomplished in the next decade. When that happens, databases such as the one at TIGR and Harvard University will contain the genomes of the vast majority of plant pathogenic bacterium, fungi, viruses, and mycoplasmas as well as the insect pests that plague agriculture around the world. Studying the plant–microbe or plant–insect interaction in the future will be done at the whole genome, transcriptome, proteome, and metabolome level of the host, and the pests and pathogens.
3.2.8 Modified plant architecture and development
Shoot architecture describes the way in which the aerial portions of plants develop. It is influenced predominantly by shoot branching and plant height. Agronomic interest in shoot architecture has stemmed mainly from potential yield increases associated with some plant ideotypes. For instance, height reduction in cereals has led to yield improvement through decreased lodging, whilst reduction of tillering in some cereals may lead to yield improvements (Duggan et al., 2005).
In sugarcane, complex interrelationships exist among shoot architecture, stalk characteristics, cane yield, and sucrose content (Milligan et al., 1990). For instance, two elements of sugarcane shoot architecture, the number and weight of stalks, determine cane yield. However, stalk number and stalk weight are negatively correlated and, thus, selecting for either trait alone may not result in increased cane yield (Milligan et al., 1990; Bell et al., 2004; Bell and Garside, 2005). Stalk architecture can also affect cane sucrose content. Taller canes are more likely to lodge under wet or windy conditions, causing a reduction in sucrose content (Singh et al., 2000, 2002), whilst clones with a propensity for producing the low-sugar-content suckers reduce the sucrose content of the harvested crop (Bonnett et al., 2001, 2004a).
Recently, several genes controlling shoot architecture traits, such as stalk height and tillering, have been described (McSteen and Leyser, 2005; Wang and Li, 2006). There is now an opportunity to test how modifying stalk characteristics with these genes will affect cane yield and sucrose content in the same sugarcane cultivar without altering other genes in the plant. Three areas of research have been identified which may lead to increased yields, accelerated biomass production, controlling lodging, and reducing suckering. The genes that are currently being used to address these research areas are the TB1 gene, which has been shown to affect tiller number and stem width in maize and rice (Doebly et al., 1997; Takeda et al., 2003), the MAX3 gene that affects axillary meristem outgrowth in rice (Zou et al., 2006) and Arabidopsis (Booker et al., 2004), and several gibberrellin oxidases that regulate stem elongation (Hedden and Phillips, 2000). Transgenic sugarcane plants have been produced and are currently undergoing extensive glasshouse and field trials to test what effects these genes produce (Pribil et al., 2007).
3.2.9 Other products
3.2.9.1 Products from phenylpropanoid pathway
The phenylpropanoid pathway is an important plant-metabolic pathway involved in the synthesis of defense-related secondary metabolites and in the production of structural-reinforcement polymers, including lignin (Matsuda et al., 2005). The phenylpropanoid pathway produces a large and diverse array of chemicals, both as intermediates and as end products. Enzyme levels in the phenylpropanoid pathway are tightly regulated (Ni et al., 1996) and flux into the phenylpropanoid pathway is controlled, at least in part, via feedback regulation of PAL, sensed through production of cinnamic acid (Blount et al., 2000). PAL is regulated both transcriptionally and post-transcriptionally (Dixon and Paiva, 1995). This is an important consideration when attempting to over-produce intermediates or end products of the phenylpropanoid pathway by attempting to increase flux through the pathway.
The MYB and bHLH group of transcription factors are involved in regulation of the phenylpropanoid, anthocyanin, and flavonoid pathways. Various branches of phenylpropanoid metabolism are regulated by branch-specific activating and repressing MYB transcription factors, sometimes dependent on bHLH (Vom Endt et al., 2002). MYB transcription factors have been identified, which regulate the first steps in the phenylpropanoid pathway.
Maeda et al. 2005 isolated carrot cDNA encoding the R2R3 type of MYB transcription factor (DcMYB1) and found that transient expression of DcMYB1 could up-regulate the PAL gene (DcPAL1) promoter activity in carrot protoplasts. Induction of DcPAL1 expression occurred 1h after DcMYB1 expression in carrot protoplasts under various stress treatments, including treatment with a fungal elicitor and by ultraviolet-B (UV-B) irradiation. When DcMYB1 expression was repressed using RNA interference, up-regulation of DcPAL1 expression was negated. It was suggested that DcMYB1 is the main regulatory factor of the DcPAL1 gene that responds to environmental cues.
Jin et al. 2000 produced an Arabidopsis line that was a mutant of the R2R3 MYB gene, AtMYB4, which showed enhanced levels of sinapate esters in its leaves. The mutant line was more tolerant of UV-B irradiation than the wild type. The increase in sinapate ester accumulation in the mutant was associated with an enhanced expression of the gene encoding cinnamate 4-hydroxylase (C4H), which appears to be the principal target of AtMYB4 and an effective rate limiting step in the synthesis of phenylpropanoids. AtMYB4 works as a repressor of target gene expression and includes a repression domain. It belongs to a novel group of plant R2R3 MYB proteins involved in transcriptional silencing (Jin et al., 2000).
The ability to significantly modify flux through the phenylpropanoid pathway would be beneficial for research targeted at producing novel compounds in sugarcane. McQualter et al. 2005 evaluated sugarcane as a production platform for p-hydroxybenzoic acid using 4-hydroxycinnamoyl-CoA hydratase/lyase (HCHL) from P. fluorescens (Gasson et al., 1998) that provides a one-enzyme pathway from a naturally occurring plant intermediate (Mayer et al., 2001). The substrate for HCHL is 4-hydroxycinnamoyl-CoA (a cytosolic phenylpropanoid intermediate). HCHL elevates p-hydroxybenzoic acid levels in plants (Mayer et al., 2001), and its expression in transgenic sugarcane led to the accumulation of p-hydroxybenzoic acid glucosal conjugates to as high as 7.3% and 1.5% DW in leaf and stem tissue, respectively, with no discernible phenotypic abnormalities (McQualter et al., 2005). However, as a result of diverting carbon away from the phenylpropanoid pathway, there was a severe reduction in leaf chlorogenic acid, subtle changes in lignin composition, as revealed by phloroglucinol staining, and an apparent compensatory up-regulation of PAL. Product accumulation in the leaves at the highest level of gene expression obtained in the study was clearly substrate limited. Increasing flux through the phenylpropanoid pathway in sugarcane by increasing the expression of the PAL gene (the first enzyme in the phenylpropanoid pathway) using a sugarcane ortholog of DcMYB1 could help overcome substrate limitation problems and enable economically viable amounts of p-hydroxybenzoic acid to be produced in transgenic sugarcane. Additionally, silencing the repressor function of a sugarcane ortholog of the AtMYB4 transcription factor could increase flux through the pathway at the C4H rate-limiting step (the second step in the phenylpropanoid pathway).
3.2.9.2 Flavonoids
Flavonoids form a large group of polyphenolic compounds produced in plants that encompass the wide variety of phytochemicals present in the human diet. Basic research, animal model, and human studies suggest flavonoid intake may reduce the risk of several age-related chronic diseases (Graf et al., 2005). Duarte-Almeida et al. 2006 identified phenolic compounds in sugarcane (S. officinarum) juice showing the predominance of flavones (apigenin, luteolin, and tricin derivatives) among flavonoids, and of hydroxycinnamic, caffeic, and sinapic acids, among phenolic acids, representing a total content of around 160mgl–1. A tricin derivative was present in the highest proportion (>10% of the total). The phenolic extract obtained from sugarcane juice showed a protective effect against in vivo MeHgCl intoxication and potent inhibition of ex vivo lipoperoxidation of rat brain homogenates, indicating a potential use for beneficial health effects and/or therapeutic applications.
A transgenic approach to enhancing flavonol content in sugarcane has not yet been attempted. However, the maize transcription factor genes LC and C1 have been expressed in transgenic tomatoes, resulting in increased flavonoid levels (Bovy et al., 2002). Tomato fruit normally contain small amounts of flavonoids in the peel, but co-expression of the LC and C1 genes enabled up-regulation of the flavonoid pathway in the flesh of the tomato fruit, a tissue that does not normally produce flavonoids. This resulted in a strong accumulation of flavonols and a modest increase in flavanones.
3.3 Economic Opportunities, Industrial Perspectives, and Political and Economic Consequences
Around the world a paradigm shift is being forced by the concern that we have reached the end of the age of inexpensive crude oil. This is being caused by a shortage of crude oil brought on by (i) the rapid industrial development of China and India (Goodstein, 2004); (ii) the political instability in the major crude oil producing regions of the world; and (iii) the fact that we are either at or very close to the peak for world oil production/reserves (Hubbert, 1956). The process of weaning the world off crude oil has begun, and the transition from a petroleum-based economy to a bio-based economy is underway. Nowhere is this more apparent than in the legislation being enacted in the European Union (EU). Manufacturers in the global automotive, packaging, electronics, and chemical industries will be responsible for the end-of-life processing of their products sold or produced in the EU. This means it will become increasingly more expensive to dispose of manufactured goods, forcing companies to recycle or reuse (End-of-Life). For instance, 100% of all electronics, most parts of automobiles, and all the packaging sold in the EU will have to be reusable or recyclable.
Clear “Vision Statements” and “Roadmaps” of the EU and the US R&D funding bodies are now in place (Europa, Plants for the future; EPOBIO; http://www.suschem.org/content.php?_document[ID]=2049&pageId=2491; United States Department of Energy Genomics, GTL roadmap; http://www.eere.energy.gov/biomass/publications.html#vision). The corporate world is also investing heavily with the largest focus on biofuels. Biorefineries are being constructed worldwide, primarily to produce ethanol. Additional investments are being made in the production of fine chemicals using fermentation technologies. The production of diverse biopolymers is an area with considerable attention. Examples include: (i) Cargill's Natureworks, bioplastics made from polylactic acid; (ii) Dupont's SoronaTM (http://www2.dupont.com/Sorona/en_US), the major component, 1,3-propanediol, is being produced in a joint venture with Tate & Lyle; and (iii) Metabolix's Natural Plastics based on PHA, which will be produced in a joint venture with Archer Daniels Midland (http://www.metabolix.com/index.html). All of these polymers begin with the simple sugars found in corn starch.
What does all this have to do with transgenic sugarcane? The Visions and Roadmaps from the EU and US, the investment in R&D by governments around the world, and by corporations in new bio-based businesses can only mean opportunity (European Union Framework Seven, United States Farm Bill, DuPont, Toyota, Cargill, Abengoa). To make sugarcane attractive for investment, such as is happening in Brazil, the technologies to engineer sugarcane must be advanced. In the future, sugarcane could be genetically engineered to grow more sugar and biomass per hectare, use less water and fertilizer, be resistant to a range of pests and diseases, produce its own bioherbicide to out-compete weeds, be able to grow on marginal land, and be able to grow in drier and cooler climates. The process of harvesting sugarcane could trigger a cascade of enzymes that would start degrading the chains of sugars that make up cellulose and hemicellulose, and modify the lignin, making the sugarcane easier to mill. This would help produce a larger pool of sugars readily accessible for the biorefineries linked to the sugarcane mills. Sugarcane could also be engineered so that a range of products other than sugars could be produced in its cells while it is growing in the field, adding additional value and further diversifying its product base.
Much of the basic research to accomplish this is already underway. The core lesson from Brazil is that it takes years or even decades to develop the knowledge base, the core skills, and the infrastructure for a bio-based economy (Lorentzen, 2006; Velho and Velho, 2006). The economic prosperity that Brazil is now enjoying because of sugarcane and its byproducts has taken them almost 40 years to develop. This prosperity gives them both the money to invest and a desire to continue this growth cycle. This creates opportunities to commercialize GM sugarcane with improved properties.
The most advanced programs for commercial release of transgenic sugarcane are in Brazil, South Africa, USA, India, China, The Philippines, and Australia. The Centro de Tecnologia Canavieira (http://www.ctc.com.br) recently announced it would be field-testing three new transgenic sugarcane varieties with 15% increase in sugar yields (http://www.inovacao.unicamp.br/english/report/news-sugaralcohol060522.shtml).
The economic prosperity of the Brazilian sugarcane industry is also being mirrored in the US maize industry. The growing global bioeconomy provides opportunity for any industry that is willing to invest in the infrastructure to produce large quantities of inexpensive sugars. Sugarcane industries are particularly well suited because they already have in place the mechanism to haul very large quantities of biomass from where it is grown to where it is processed. The sugars locked up in this biomass have the interest of governments and industries alike. Many believe that the sugars in the cellulose and hemicellulosic biomass are the replacement for crude oil. The United States and the EU see the tools of the biotechnology revolution as fundamental to this process.
Biology and engineering are merging at the computer interface through the fields of genomics (Sanford et al., 2002; Morgante, 2006), transcriptomics, proteomics (Heazlewood and Millar, 2003), metabolomics (Oksman-Caldentey and Saito, 2005; Rockfort, 2005), and fluxomics (Sanford et al., 2002), generating enormous data sets that are allowing for the development of systems biology (Lee et al., 2005; Barrett et al., 2006; Joyce and Palsson, 2006). Using these databases, experiments are designed and tested on a computer before being trialed in a laboratory (Lee et al., 2005). One person working in the corner of one laboratory can accomplish more in a few months than was previously accomplished by research teams working throughout their entire careers. These powerful tools could not have come at a better time, as the rising demand for crude oil starts to impact on price, which potentially could play havoc with the world's economies (Goodstein, 2004).
3.3.1 Human health and environment
There is no reason to assume that genetic engineering has a greater potential to harm human health or the environment than any techniques used in classical breeding (Miller, 2007). If there is a lesson we have learned from the enormous experience of commercially growing GM crops on over 1 billion acres (400 million hectares) around the world for over two decades is that if the trait(s) encoded by the transgenes are safe, then there are more than sufficient safeguards in traditional breeding programs to ensure that the GM-derived crop is safe (Bradford et al., 2005).
However, there is still considerable resistance to food derived from GM plants, even though there is no scientific basis for this concern (Miller, 2007). Because of this resistance, the large buyers of raw sugar have been requiring guarantees from the suppliers that the sugar supply is not derived from GM plants. This has created a wait-and-see approach, where sugarcane biotech research and development is ongoing but no one wants to be the first into the market. However, it is likely that sugar from sugarbeets engineered with herbicide and pest resistance traits will enter the market first and that will hopefully open the door for sugarcane. If GM sugarcane lines with doubled sugar production (Wu and Birch, 2007) perform well in field trials, pilot-scale production will most likely be trialed.
Refined white sugar (>99.98% sucrose) is the most chemically pure food produced from agriculture, thus, reducing any foundation for concern about foreign genes or gene products in this food (Birch and Maretzki, 1993). Taylor et al. 1999 analyzed the various products during the crystallization process of sugar from sugarcane juice for the presence of transgene DNA. Their results showed that no PCR amplifiable DNA was present in sugar crystallized from SCMV transgenic plants.
However, when sugarcane is engineered as a biofactory to produce fine chemicals or pharmaceuticals, it is critical that clear safeguards are in place to ensure that there is no conceivable harm to public health or the natural environment. Safety evaluations will have to be done on a case-by-case basis. For instance, sugarcane producing bioplastic such as the pHBA and PHAs discussed in this chapter could still be used for production of sugars for human consumption. Animals already make pHBA and have the capacity to digest it (McQualter et al., 2005), and PHAs have a 40-year history of safety in a variety of medical applications, especially because of biodegradability and biocompatibility. PHAs have been used for surgical sutures and wound dressings. There is ongoing research for the commercialization of PHA-based products for tissue engineering and for controlled release systems, and for bone repair (Zhijiang, 2006). Because these biopolymers in nature are used as a carbon sinks, they are fully biodegradable. So producing them in sugarcane should not be dangerous to human health or the environment.
3.4 Conclusions
Since the first successful report of transgenic sugarcane plants less than 20 years ago, biotechnology has advanced rapidly and been adopted by sugar industries and research organizations worldwide. Research into a range of input traits such as pest and disease resistance, sugar quality and shoot architecture, and output traits such as increased sucrose levels, alternative sugars, biopolymers, pharmaceuticals, and bulk proteins are ongoing. The future looks bright for sugarcane, as market forces drive the development of the bioeconomy, creating opportunities for sugar industry diversification and new partnerships with other global and local industries. A combination of factors, including but not limited to, global warming, instability in the Middle East, concern over long term oil supplies, and the industrialization of China and India, have caused the world to look for alternatives to petroleum for fuel, energy, and fine chemicals. Governments have made plans, drawn up roadmaps, and set priorities for R&D funding to create ways to shift from a disproportional dependence on the nonrenewable resource petroleum. They are creating new industries and whole new economies based on renewable sources. The sugar derived from lignocellulosic biomass is going to be the renewable feedstock for the bioeconomy. The infrastructure is already being developed. Biorefineries are being constructed all over the United States, Europe and Brazil. Ethanol and a range of biopolymers are being produced in these biorefineries. In the United States, maize is the feedstock; in Brazil it is sugarcane. These nations all have R&D programs underway to switch over to biomass.
Developing new cultivars of sugarcane that will produce more tonnage, use less water and fertilizer, that will not fall down in a storm, can tolerate floods or drought, grow in the heat or the cold, are resistant to pests and diseases, can tolerate salt, can fight off weeds, and supply the majority of the world's needs for sucrose, and other sugars, bioplastics, and a host of other fine chemicals, reduce greenhouse gas, and clean up the environment is going to require considerable genetic manipulation. Systems biotechnology will help in the design strategies to manipulate the metabolic pathways of sugarcane. However, the ability to transform sugarcane with genes and/or metabolic pathways will be instrumental in helping sugarcane industries become drivers of the bioeconomy.
4 Acknowledgments
Our special thanks to Dr Peter Allsopp from BSES Limited for his editing and advice on this book chapter. We also thank Drs Debra Stenzel (Analytical Electron Microscopy Facility, Queensland University of Technology, Australia) and Lars Petrasovits (Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Australia) for their assistance with the electron micrographs in Figure 5. Annathurai Gnanasambandam is a recipient of a “Smart State Fellowship” awarded by the Department of State Development, Trade, and Innovation of the Queensland Government.
- ADM
- Archer Daniels Midland
- AFLP
- amplified fragment length polymorphisms
- AG
- AGAMOUS
- AP1
- APETALA1
- Adh
- alcohol dehydrogenase
- BAC
- bacterial artificial chromosome
- BC1
- backcross 1
- BSV
- banana streak virus
- CMR
- Comprehensive Microbial Resource
- CSP
- Community Sequencing Program
- CTC
- Centro de Tecnologia Canavieira
- CaMV
- cauliflower mosaic virus
- DCL
- defective chloroplast and leaves
- DW
- dry weight
- ER
- endoplasmic reticulum
- ESTs
- Expressed sequence tags
- ESTs
- Expressed sequence tags
- EU
- European Union
- EU
- European Union
- FDV
- Fiji disease virus
- G6P
- D-glucose-6-phosphate
- GFP
- green fluorescent protein
- GM
- genetically modified
- GM-CSF
- human cytokine granulocyte macrophage colony stimulating factor
- HCHL
- 4-hydroxycinnamoyl-CoA hydratase/lyase
- JGI
- Joint Genome Institute
- LCB
- lesser corn-stalk borer
- LDNA
- linearized plasmid DNA
- LFY
- LEAFY
- MARS
- matrix attachment regions
- MITE
- miniature inverted-repeat transposable element
- MS
- Murashige and Skoog
- OCS
- octopine synthase
- ONSA
- Organization for Nucleotide Sequencing and Analysis
- ORF
- open reading frame
- Ocs
- octopine synthase gene
- PAL
- phenylalanine ammonia-lyase
- PCR
- polymerase chain reaction
- PEG
- polyethylene glycol
- PFP
- Pyrophosphate-dependent phosphofructokinase
- PHA
- polyhydroxyalkanoate
- PHB
- polyhydroxybutyrate
- PIG
- particle inflow gun
- PMI
- phosphomannose isomerase
- PPO
- polyphenol oxidase
- PTGS
- post-transcriptional gene silencing
- QTL
- quantitative trait loci
- RGS
- G-protein signalling protein
- RLU
- relative light units
- RNAi
- RNA interference
- ROS
- reactive oxygen species
- S6P
- sorbitol-6-phosphate
- S6PDH
- sorbitol-6-phosphate dehydrogenase
- SCBV
- sugarcane bacilliform virus
- SCMV
- Sugarcane mosaic virus
- ScYLV
- sugarcane yellow leaf virus
- SoGI
- Saccharum officinarum Gene Index
- SrMV
- Sorghum mosaic virus
- TIGR
- Institute for Genomic Research's
- T-DNA
- transfer DNA
- UDP
- uridine diphosphate
- USDA
- US Department of Agriculture
- UV-B
- ultraviolet-B
- VIGS
- Virus induced gene silencing
- acetyl-CoA
- acetyl-coenzyme A
- albD
- albicidin detoxification gene
- cDNA
- complementary DNA
- dsRNA
- double-stranded RNA
- hph
- hygromycin phosphotransferase
- mRNA
- messenger-RNA
- mds6pdh
- M. domestica sorbitol-6-phosphate dehydrogenase gene
- pHBA
- p-hydroxybenzoic acid
- pHBA
- p-hydrobenzoic acid
- pHBA
- p-hydroxybenzoic acid
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative reverse-transcriptase polymerase chain reaction



