Genetic control of collagen-induced arthritis in a cross with NOD and C57BL/10 mice is dependent on gene regions encoding complement factor 5 and FcγRIIb and is not associated with loci controlling diabetes
Abstract
The nonobese diabetic (NOD) mouse spontaneously develops autoimmune-mediated diseases such as diabetes and Sjögren′s syndrome. To investigate whether NOD genes also promote autoimmune-mediatedarthritis we established a NOD strain with an MHC class II fragment containing the Aq class II gene predisposing for collagen induced arthritis (NOD.Q). However, this mouse was resistant to arthritis in contrast to other Aq expressing strains such as B10.Q and DBA/1. To determine the major resistance factor/s, a genetic analysis was performed. (NOD.Q×B10.Q)F1 mice were resistant, whereas 27% of the (NOD.Q×B10.Q)F2 mice developed severe arthritis. Genetic mapping of 353 F2 mice revealed two loci associated with arthritis. One locus was found on chromosome 2 (LOD score 9.8), at the location of the complement factor 5 (C5) gene. The susceptibility allele was from B10.Q, which contains a productive C5 encoding gene in contrast to NOD.Q. The other significant locus was found on chromosome 1 (LOD score 5.6) close to the Fc-gamma receptor IIb gene, where NOD carried the susceptible allele. An interaction between the two loci was observed, indicating that they operate on the same or on interacting pathways. The genetic control of arthritis is unique in comparison to diabetes, since none of these loci have been identified in analysis of diabetes susceptibility.
Abbreviations:
-
- CIA:
-
Type II collagen-induced arthritis
-
- CII:
-
Type II collagen
-
- RCII:
-
Rat type II collagen
-
- RA:
-
Rheumatoid arthritis
-
- C5:
-
Complement factor 5
-
- NOD:
-
Nonobese diabetic
1 Introduction
The development of autoimmune diseases, such as rheumatoid arthritis (RA) and type I diabetes, is complex and dependent on multiple genes and environmental factors. Each of these disease entities may also be caused by different pathogenic pathways in different individuals. Animal models are useful for elucidating the genetic control of the various pathways leading to disease. The nonobese diabetic (NOD) mouse strain, which spontaneously develops a disease mimicking type I diabetes, has been useful in such analysis 1. The major gene regions controlling developmentof diabetes have been identified mainly from crosses with the C57BL/10 (B10) strain 1, 2. Not surprisingly, a major locus was found on chromosome 17 that contains the MHC region 3, 4. The H2g7 haplotype of the NOD mouse promotes development of diabetes whereas other haplotypes have a dominant protective effect. These genes have not been defined yet but it is believed that one of them is the class II A gene, although neither its peptide binding specificity nor its function in diabetes development are known.In addition to the MHC region, at least 16 different loci have been suggested to be linked to diabetes (Fig. 1) 1–3, 5. The underlying genes have not been determined, although a role for the IL2 gene on chromosome 3 6 and the co-stimulatory CTLA-4 and CD28-encoding genes on chromosome 1 7 have been proposed to be of importance. These genes relate to T cell expansion and activation and there is strong evidence that T cell-mediated pathways are of importance for development of diabetes.
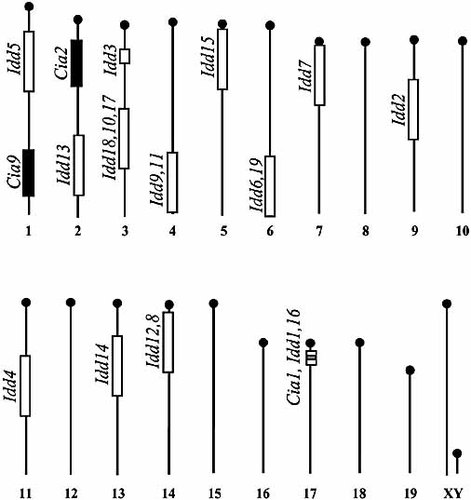
The NOD-associated arthritis (Cia) and diabetes (Idd) loci are located at different regions except for the MHC. The figure shows the reported diabetes loci in the NOD mouse together with the Cia2 and Cia9 loci described here 2, 3, 5, 48–54. The previously defined Cia1 (the MHC region) did not come up in the F2 intercross since NOD.Q and B10.Q have the same MHC haplotype H2q 8.
In the present study we have asked the question whether NOD genes also control the development of other autoimmune-mediated inflammatory processes such as arthritis. Collagen-induced arthritis (CIA), an established model for RA, can be induced in mice by immunization with type II collagen (CII) 8. Susceptibility is controlled by specific MHC class II genes, such as Aq, that binds defined immunodominant CII peptides 9. Activation of CII peptide specific T cells trigger an immune-mediated cascade resulting in an inflammatory destruction of the peripheral joints 10. Many pathways and factors are likely to participate, illustrated by the fact that mice lacking B cells, T cells, complement factor 5 (C5) or the FcRγ chain are resistant to disease induction 11–14.
To determine the contribution of NOD genes to CIA, we first made a NOD strain congenic for the H2q haplotype, NOD.Q mice. The NOD.Q mice were found to be resistant to CIA. Analysis of an F2 intercross showed that the major influence on arthritis was exerted by a region containing the gene encoding C5 on chromosome 2 and a region containing a polymorphic FcγRIIb encoding gene (Fcgr2b) on chromosome 1. Surprisingly, neither of these loci have been found to be associated with diabetes.
2 Results
2.1 H2q congenic NOD mice are resistant to collagen induced arthritis
An H2q NOD strain was established after ten backcrosses of a C3H.Q derived fragment into the NOD background. The fragment was found to be 4.5 cM around the MHC on chromosome 17 (Fig. 2). Susceptibility to CIA after immunization with heterologous CII is strongly associated with the H2q haplotype. However, the NOD.Q strain was resistant to CIA in contrast to other H2q strains (DBA/1, B10.Q, C3H.Q) 15 (Table 1, Fig. 3). No diabetes, but sialadenitis and mild insulitis were observed in the NOD.Q strain (data not shown).
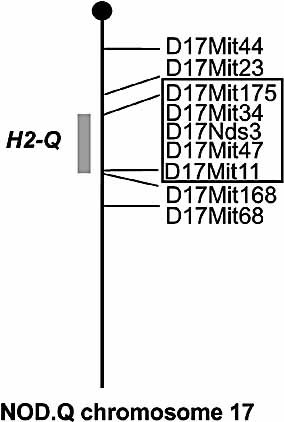
The MHC fragment on chromosome 17 in the NOD.Q mouse. The box indicates the borders of the inserted fragment.
The B10.Q strain was chosen for further comparative experiments with the resistant NOD.Q strain. The B10.Q strain developed arthritis of intermediate severity with approximately 50% incidence, which is in accordance with earlier experiments (Table 1). Interestingly, H2q NOD congenic (NOD.Q) mice developed no signs of arthritis (Table 1). The NOD genetic background dominantly suppressed the disease development as shown by an F1 intercross between NOD.Q and the B10.Q where no signs of arthritis could be detected (Table 1, Fig. 3).
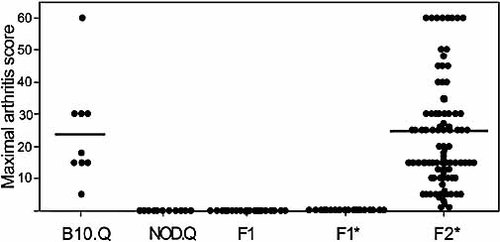
Arthritis severity. The maximal arthritis score, as a measurement of disease severity, for individual B10.Q, F1 and F2 mice. Only arthritic mice were calculated. The mean value is marked as a line. *One group of the F1 mice and all the F2 mice were given an i.p. injection of pristane 2 weeks before immunization.
|
Strain |
n |
Incidence (%) |
Mean day of arthritis onsetb) |
Mean max clinical scoreb, c) |
|---|---|---|---|---|
|
B10.Q |
22 |
46 |
46 ± 20 |
24 ± 15 |
|
NOD.Q |
11 |
0 |
− |
− |
|
(NOD.Q × B10.Q)F1 |
20 |
0 |
− |
− |
|
(NOD.Q × B10.Q)F1a) |
20 |
0 |
− |
− |
|
(NOD.Q × B10.Q)F2a) |
353 |
27 |
44 ± 15 |
25 ± 17 |
- a) Some F1 mice and all the F2 mice were given pristane i. p., 2 weeks before immunization with RCII and Freund's complete adjuvant.
- b) Mean day of arthritis onset and mean max clinical score are presented as mean ± SD.
- c) Only diseased animals were included with the mean maximal score.
2.2 Analysis of an F2 cross of NOD.Q and B10.Q strains
To further analyze the cause of the resistance of NOD.Q as compared with B10.Q we produced 353 F2 intercross mice that were immunized with rat type II collagen (RCII) in CFA to induce CIA. The (NOD.Q × B10.Q) F1 did not develop arthritis, so it could be suspected that the incidence of arthritis in the F2 intercross would be quite low, and therefore the F2 mice were given an i.p. injection of pristane 2 weeks before the immunization in an attempt to enhance the incidence of CIA 16. An i.p. injection of pristane 2 weeks prior to immunization with RCII in CFA did not induce arthritis in F1 (n=20) animals (Table 1 and Fig. 3). The mice were examined for arthritis until day 110. Serum was obtained on day 35 and analyzed for the levels of C5.
The mice started to develop arthritis 4 weeks after immunization. The frequency of arthritis was 27% and the day of onset, duration and degree of severity were highly variable between individual mice. The disease traits analyzed were day of onset of arthritis and maximal arthritis score.
The F2 mice could be divided in different groups based on the C5 serum levels, a pattern that was expected since the NOD strain is known to be completely deficient in C5 caused by a disrupted gene encoding C5 17 (Fig. 4).
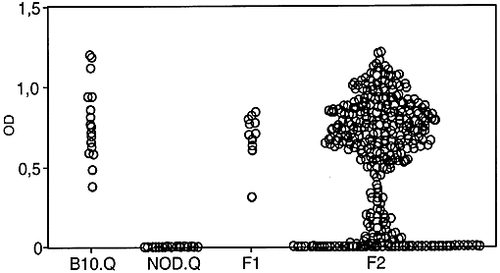
C5 levels in sera. Sera were taken 35 days after immunization and assayed for levels of C5. The figure shows the C5 levels in sera of B10.Q, NOD.Q, F1 and F2 mice.
The F2 mice were mapped using 155 markers with an 8.6 cM inter-marker distance (based on calculations using MAPMAKER) covering the complete genome. Linkage to arthritis severity as well as to the serum levels of C5 were analyzed and the results summarized in Table 2. No differences in the linkages reported here were observed between males and females (data not shown).
|
QTL |
Affected phenotype |
Chromosome |
Associated microsatellite marker |
LOD score |
Inheritance pattern |
Explained variance (%) |
|---|---|---|---|---|---|---|
|
Cia2 |
Arthritis severity |
2 |
D2Mit7 |
9.8 |
B10.Q additive |
12 |
|
Cia2 |
C5 |
2 |
D2Mit32 + 6 cM |
B10.Q dominant |
77 |
|
|
Cia9 |
Arthritis severity |
1 |
D1Mit146, D1Mit270 |
5.6 |
NOD.Q additive |
7 |
|
– |
Onset of arthritisa) |
− |
− |
− |
− |
− |
- a) No linkage to onset was found.
2.3 The resistance to CIA is mainly caused by C5 deficiency
The C5 deficiency was controlled by a locus on chromosome 2 (LOD score 83.5), at the location of the C5 encoding gene (Fig. 5A), known to be deficient in the NOD strain 17. Thus, the levels of C5 in serum were dominantly controlled by the B10.Q allele with no influence of heterozygosity.
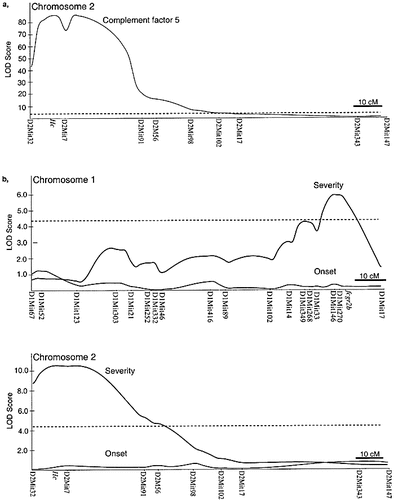
LOD score curves on chromosome 1 and 2 for linkage to (a) C5 and (b) arthritis severity and onset of arthritis. The dotted line indicates the significance level of 4.3.
For the disease severity trait, two genetic regions contained significant linkages. On chromosome 2 a linkage was found with a LOD score of 9.8. The susceptible B10.Q allele was additive. The C5 encoding gene is located at the peak of the LOD score curve and the linkage of C5 levels in serum was highly significant for markers surrounding the C5 gene (Fig. 5). However, four C5-deficient mice developed arthritis (as shown by the serum analysis of C5), showing that C5 sufficiency was not a complete requirement for development of CIA. The C5 levels were similar between heterozygous and B10.Q homozygous mice (Fig. 4).
2.4 The NOD background contributes with the disease promoting FcγRIIb encoding allele
Another locus with a significant linkage to disease severity was found on chromosome 1 with a peak LOD score of 5.6 at the location of the Fcgr2b gene (Fig. 5B). In this case the NOD conferred the susceptibility allele with an additive inheritance. This locus on chromosome 1 was denoted Cia9. No linkage was found to the day of onset of arthritis (Fig. 5B and C).
2.5 The Cia2 and Cia9 loci interact
Interestingly, heterozygosity at the Cia2 locus had different effects dependent on the allele at the Cia9 locus. Thus, mice were resistant if they were homozygous for the B10 allele at Cia9, but were susceptible if they had the NOD allele at Cia9 (p value <0.05) (Fig. 6). It is therefore likely that the presence of a NOD allele at Cia9 promotes the same pathway as the B10 allele at Cia2.
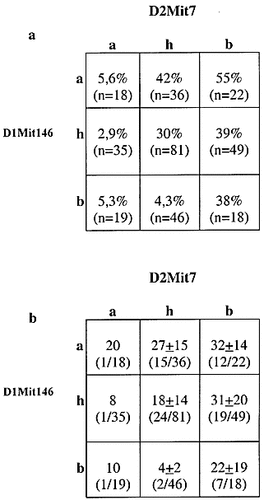
Interaction of the arthritis severity controlling loci on chromosome 1 and chromosome 2. The interaction of the loci on chromosome 1 and chromosome 2 are shown for (a) the development of arthritis reported as incidence, (b) arthritis severity reported as the mean maximal score of the individual diseased animals ± SD. The haplotypes were defined as follows; a = NOD.Q; b = B10.Q; h =heterozygous.
To confirm this interaction between Cia2 and Cia9, we created congenic mice containing the NOD allele at the Cia2 locus on a B10.Q background using the speed congeneic technique. We denoted these mice B10.Q.Cia2. To test if the interaction between Cia2 and Cia9 could be reproduced, B10.Q.Cia2+/– (n5) and their wild-type littermates were immunized with RCII in CFA to induce CIA.
The B10.Q.Cia2+/– mice (heterozygous for the Cia2 and containing the B10 allele at Cia9) showed a lower incidence of CIA and a significantly less severe disease (p <0.0001) (Fig. 7). These results confirmed the observed interactions in the F2 cross and showed that the effect of the heterozygous Cia2 locus was significant in an environment where the Cia9 locus was of B10.Q origin.
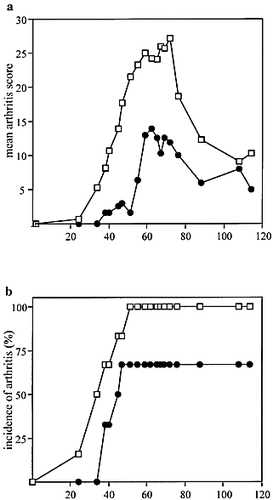
The interaction between Cia2 and Cia9 was confirmed in B10.Q.Cia2 congenic mice. Five B10.Q.Cia2+/- mice (heterozygous for the Cia2 and the B10 allele at Cia9), filled circles, and five wild-type littermates (B10.Q), open squares, were immunized with RCII in CFA to induce CIA and boosted at day 35 with RCII in incomplete Freund's adjuvant. The mice were investigated for arthritis for 120 days. The severity of arthritis (a) and incidence of arthritis (b) are shown.
3 Discussion
The NOD mouse background, which promotes several autoimmune and chronic inflammatory disorders such as insulitis and sialadenitis, was found to be highly resistant to collagen-induced arthritis. A gene segregation experiment in which the CIA MHC congenic NOD.Q strain was crossed with the B10.Q strain, then intercrossed, identified two major genetic regions controlling the disease, neither of which have previously been described to be of importance for diabetes (Fig. 1 and Table 2).
Clearly, CIA is controlled by genes other than those associated with diabetes. A major locus is Cia2 13 on chromosome 2 that contains the C5 encoding gene. The C5 deficiency of the NOD strain does not prevent development of diabetes and there is no linkage to this region in various crosses of NOD mice with C5 sufficient strains, indicating that C5 has no impact on the development of diabetes. In CIA, on the other hand, the role of the gene encoding C5 has been demonstrated by several investigators 13, 18–21. Mouse strains lacking C5 are relatively resistant to CIA 22 and gene segregation experiments have indicated chromosome 2 and C5 deficiency to be of importance 13, 23. In addition, treatment with anti-C5 neutralizing antibodies efficiently blocks the development of arthritis 21. These data, taken together, support an important role of C5 in the development of CIA. However, this does not necessarily mean that C5 is an absolute requirement for arthritis. In fact, in our experiment a few C5-deficient mice developedarthritis. This is in accordance with our earlier finding that the presence of C5 did not reach a significant linkage in a cross involving the highly susceptible DBA/1 strain in which the disease was induced with a single immunization without booster 24. Possibly, the C5-deficient mice develop arthritis along other pathways than the mice not lacking C5. However, the number of these mice in the present study was too low for elucidating which loci control the arthritis in this situation. Instead, we found a locus on chromosome 1 in which the NOD allele had an additive effect to promote arthritis. We have denoted this newly identified QTL Cia9 to distinguish it from earlier identified loci shown to be associated with CIA in various mouse crosses 23, 25, 26. This region on chromosome 1 has not been shown to be of importance for diabetes but in different lupus models 27–30, indicating again that different autoimmune models are not controlled by the same set of genes. The linkage peaks at the location of the Fcgr2b gene. The NOD mouse has a haplotype in this region that is shared with many lupus-prone strains but also with 129/sv and 129/Ola mouse strains and harbors several polymorphic genes of potential importance for autoimmune disease 27, 29, 30. One of these genes could be the Fcgr2b gene itself which is polymorphic between the NOD and B10 strains and which encodes an inhibitory immune complex-binding receptor 31. In NOD mice the FcγRIIb expression is severely reduced on macrophages and moderately reduced on activated B cells and wouldbe a likely explanation for the observed effect. In fact, deletion of the Fcgr2b gene backcrossed into the DBA/1 (H2q) mouse leads to a dramatically more severe development of CIA 14 and also contributes to increased susceptibility to other autoimmune diseases such as gastritis. However, there are several other candidate genes in the region such as Fcgr3 and Fcer1 and at least 3 other genes of importance for lupus 28, 29, 32. It is likely that this haplotype contains a cluster of naturally selected polymorphic genes of wide importance for many autoimmune diseases. These genes are likely to cooperate along various disease pathways of critical importance in order to identify the basis for autoimmune pathology.
One interesting possibility is that the presently identified genetic regions on chromosomes 1 and 2 in fact operate along the same or interacting pathways. The FcγRIIb is an inhibitory immune complex-binding receptor 33. The deletion of the Fcgr2b gene render mice highly susceptible for CIA and they develop high levels of antibodies against collagen type II 14, indicating a role for FcγRIIb in the regulation of arthritogenic antibodies in CIA. It should be noted, however, that this finding does not confirm the role of the Fcgr2b gene, nor exclude other nearby genes on chromosome 1, since the deleted gene is located in a fragment derived from the 129/Sv mouse carrying the same haplotype as the NOD mice. It has also been reported that C5 is an absolute requirement for the induction of arthritis via injection of anti-collagen type II antibodies 19. Further evidence showing that the Cia2 and Cia9 loci are in fact playing a role along the same pathway leading to arthritis is given here. Thus, we observed that in mice with a resistant Cia9 allele (B10 homozygous) there was a significant difference in arthritis incidence between Cia2 heterozygous compared to Cia2 B10 homozygous mice in the F2 cross. These data were confirmed in Cia2 congenic mice, as the B10.Q.Cia2 heterozygous mice had a lower incidence of arthritis and a less severe disease than the wild-type littermates. Although we did not find any differences in the C5 levels insera between mice with the B10 haplotype and heterozygous at the Cia2 locus it does not exclude a lower level of C5 in the joint that exert an protective effect together with the B10 allele of the Fcgr2b gene at Cia9 locus. These data argue for that an immune complex dependent pathway is important for the development of severe collagen induced arthritis. Such a finding is compatible with previous findings in the CIA model induced with heterologous CII. Development of CIA is critically dependent on B cells since B cell-deficient mice are completely resistant to disease 11 and because arthritis can be transferred with antibodies reactive with cartilage in vivo and specific for epitopes on CII 10, 34–37. The pathogenic antibodies are IgG, produced by B cells triggered through interactions with T cells specific for a heterologous epitope on CII. The binding of antibodies on cartilage may trigger macrophages and attract neutrophils through both complement and Fc receptor-dependent pathways. A direct role for T cells that not only trigger the B cells but also interact in the effector mechanisms in this immune complex-mediated pathway is also possible.
However, it is likely that also other pathways are important for development of CIA. In several other crosses of both mice and rats there is so far no evidence for a role of complement factors, or for the various Fc receptors identified in the control of the disease 25, 26, 38–41. In the present combination of mice, the NOD.Q and B10.Q, the identified loci do not explain the complete resistance in the F1 animals. It is likely that the resistance is explained by other genes, which do not show significant linkage in an environment with the strong effect of the loci on chromosome 2 and 1. A backcross of F1 with the susceptible B10.Q mouse is likely to reveal loci that may control other pathways than the ones that are controlled by the Cia2 and Cia9 loci.
Furthermore, it is likely that other autoimmune diseases, such as diabetes and Sjögren′s syndrome do not use the same type of immune complex mediated pathway as the CIA model, possibly becausea direct role of pathogenic antibodies is not as important in these diseases as in CIA.
4 Materials and methods
4.1 Mice
The H2q congenic NOD strain (NOD.Q) was made by crossing C3H.Q mice (H2q) (origin D.C. Shreffler, St. Louis, MO) and NOD mice (H2g7) (origin Jackson Laboratories, Bar Harbor, ME). Females from each F1 cross were backcrossed to males from the NOD parental strain for ten generations. This was followed by three intercrosses to obtain H2q homozygous (NOD.Q+/+) mice. The C57BL/10 (B10.Q) MHC congenic strain originated from Prof. Jan Klein, Tübingen, Germany. The B10.Q, NOD.Q, (NOD.Q × B10.Q)F1 and (NOD.Q × B10.Q)F2 mice were bred, kept and used in the animal house of Medical Inflammation Research unit, Lund University. The mice were kept in a climate-controlled environment with 12-h light/dark cycles, housed in polystyrene cages containing wood shavings, fed with standard rodent chow and water ad libitum in a specific pathogen-free environment (as defined in http://net.inflam.lu.se/).
The B10.Q.Cia2 congenic strain was constructed with the speed congenic technique 42. NOD.Q was backcrossed to B10.Q for five generations (5N). The N5 animals had "contaminating" NOD genome left on the background genome varying from around 0 to 6%. The congenic fragment on chromosome 2 varied from around 17–66 cM (the smallest ranging from marker Hc to D2Mit91 and the largest ranging from D2Mit296 to D2Mit343). The animals were considered heterozygous when they were heterozygous for the C5 marker, Hc.
4.2 Antigens
RCII was prepared from Swarm chondrosarcoma as earlier described 43. The collagen was pepsin digested and further purified and finally dissolved in 0.1 M acetic acid and stored at 4°C until usage.
4.3 Induction of CIA
Mice (male and female), 8–16 weeks of age, were immunized at the base of the tail with 100 μg RCII emulsified 1:1 in Freund's complete adjuvant (Difco, Detroit). Boost injections with halfthe dose RCII and emulsified in Freund's incomplete adjuvant (Difco) were given at day 35 post immunization. The (NOD.Q × B10.Q)F2 and some F1 mice were given an i.p. injection with 100 μ lpristane, 2 weeks before the immunization with RCII in an attempt to enhance the incidence of arthritis. Arthritis was evaluated blindly using a scoring system based on the number of inflamed joints in each paw, inflammation being defined by swelling and redness. In this scoring system, described in detail elsewhere 38, each inflamed toe or knuckle gives 1 point, whereas aninflamed wrist or ankle gives 5 points, resulting in a score of 0–15 (5 toes + 5 knuckles + 1 wrist/ankle) for each paw and 0–60 points for each mouse.
4.4 Measurement of anti-collagen type II IgG levels
Mice in the arthritis experiments were bled at day 35 after immunization and the level of mouse anti-RCII IgG antibodies (μg/ml) were measured in the sera, using ELISA technique as previously described 44.
4.5 Detection of complement factor 5 in sera
The levels of C5 in sera were detected using the ELISA technique as previously described 45. Briefly, sera taken 35 days after immunization were added to 96-well plates (Costar, Corning Incorporated, Corning, NY) coated with a chicken anti-mouse C5 antibody. To detect C5 levels, a biotinylated chicken anti-mouse C5 antibody were used followed by a streptavidin alkalinephosphatase (Jackson Immuno Research, West Grove, PA) incubation. Paranitrophenol was used as a chromogenic substrate and the absorbance determined in a Titertek multiscan spectrophotometer.
4.6 Genotyping and linkage analysis
Microsatellite markers were purchased from Interactiva Biotechnology Inc (Ulm, Germany). The order of the markers was based on the map available from the Jackson laboratory (www.jax.org). A full genomic scan was performed on 97 animals using 155 microsatellite markers, with an average inter-marker distance of 8.6 cM. The complete map is available on http://net.inflam.lu.se/. Another 95 animals were scanned from chromosome 1–14 using 120 microsatellite markers. For regions where the LOD scores exceeded 1.5, the remaining 161 animals were typed using 85 microsatellite markers.
All markers were assayed by PCR as follows: Genomic DNA (15 ng) was amplified in a final volume of 9 μl containing dNTP (200 M), MgCl2 (1.5 mM), primer (1.5 pmol, of each) and Taq GOLD DNA polymerase (0.2 U) (Perkin Elmer Biosystems). The forward primer was labeled with a fluorescent dye. Amplification conditions were as follows: 95°C for 1.1 min, followed by 33 cycles of 95°C for 30 s, 55°C for 75 s, 72°C for 75 s and a final extension at 72°C for 7 min. The reactions were performed using an ABI 877 Integrated Thermal cycler (Applied Biosystems, Inc., Foster City, CA), followed by an automatic pooling of the products. The PCR products were size-fractionated on 4% polyacrylamide gels, on an ABI 377 sequencer and the sizes of the fragments were determinedusing the Genescan software version 3.1 (Applied Biosystems, Inc.) with TAMRA GS-350 or 500 as internal size standard.
Linkage analysis was performed using MAPMAKER 46, 47. Linkage was tested using as phenotypes maximal score of arthritis, onset of arthritis and the level complement factor 5 in sera.
The Mann-Whitney U-test was used for comparisons between groups. Differences were regarded as significant if the p value was < 0.05. The differences in arthritis severity betweenthe B10.Q.Cia2 heterozygous mice and B10.Q wild-type littermates was calculated as the difference in the area under the severity curve.
Acknowledgements
We wish to thank Anders Larsson at Uppsala University for kindly providing antibodies for the complement factor 5 assay. Inger Jonasson, Ann-Sofi Strand andFredrik Heeder for help with the genotyping. Lennart Lindström, Carlos Palestro and Marina Persson for taking good care of the animals. This work was supported by grants from the Anna Greta Crafoord Foundation for Rheumatological Research, King Gustaf V: s 80-year foundation, the Kock and Österlund Foundations, the Swedish Association Against Rheumatism, the Swedish Medical Research Council, the European Union BIOMED program, the Research Council of Norway (115563/320), the Norwegian Foundation of Health and Rehabilitation and the Broegelmann Foundation.
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH




