Cloning and sequence analysis of the Candida boidinii ADE2 gene
Abstract
Candida boidinii ADE2 gene (phosphoribosyl-5-aminoimidazole carboxylase; AIRC, EC 4. 1. 1. 21) has been cloned by homology to the Saccharomyces cerevisiae ADE2 gene. The cloned C. boidinii ADE2 gene complemented the ade2 mutation of S. cerevisiae. Sequence analysis showed a single open reading frame of 1719 nucleotides coding for a polypeptide of 573 residues. Comparison of the deduced amino acid sequence with those of AIRC enzymes from other yeasts showed marked homology among yeast AIRCs. The sequence has already been submitted to DDBJ/EBML/GenBank under Accession No. AB034950. Copyright © 2000 John Wiley & Sons, Ltd.
Introduction
Recently, methylotrophic yeasts Candida boidinii, Pichia pastoris and Hansenula polymorpha have been developed as advantageous yeast expression systems (Faber et al., 1995; Cregg et al., 1993; Sakai et al., 1995). The methylotrophic yeast expression system has several merits, including: (a) cells can be grown to high density on a low-cost medium because methanol is an inexpensive and non-fermentable carbon source; (b) methylotrophic yeasts have strong and tightly regulated promoters (e.g. alcohol oxidase promoters).
The asporogenous yeast C. boidinii has been thoroughly investigated biochemically as a model organism for peroxisome assembly (Goodman et al., 1990) or industrial production of metabolites from methanol (Tani, 1991). However, because of its inability to form spores, breeding of C. boidinii had been limited to conventional mutagenesis or gene disruption. The ADE2 gene, encoding phosphoribosyl-5-aminoimidazole carboxylase (AIRC) is a useful selectable marker for the yeast transformation system. Lack of AIRC activity leads to accumulation of a red pigment. Consequently, colonies of ade2 mutants are usually red in colour, in contrast to the white-coloured wild-type colonies. This phenotype is useful as a visual signal for differentiating Ade− from Ade+ strains when the gene is used as a marker of transformation or recombination. In this report, we performed cloning and sequence analyses of the C. boidinii ADE2 gene that could be employed as a useful selectable marker.
Materials and methods
Strains and media
C. boidinii KST25 was used as the source of genomic DNA library. C. boidinii KST2515 (ura3) strain was obtained from C. boidinii KST25 according to the method described previously (Sakai et al., 1991, 1992b). Saccharomyces cerevisiae S288C (MATa SUC2 mal mel gal2 CUP1) was used to isolate the ADE2 gene, and YPH499 (MATa ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1) was used as a host for transformation to examine the function of cloned genes. Yeast cells were cultured at 30°C in YPD medium (1% Bacto-yeast extract, 2% Bacto-peptone, and 2% glucose) or minimal medium. Minimal medium contained 0.67% Yeast Nitrogen Base without amino acids, 2% glucose, and amino acids adenine and uracil added according to strain requirements (Sherman et al., 1986). Escherichia coli DH5α (Sambrook et al., 1989) was used for plasmid construction. E. coli cells were cultured in 2×YT medium (1.6% Bacto-tryptone, 1.0% Bacto-yeast extract, and 0.5% NaCl), supplemented with ampicillin at 100 µg/ml.
DNA methods
Recombinant DNA methods were performed as described in Sambrook et al. (1989). The S. cerevisiae ADE2 coding sequence (Stotz and Linder, 1990) was obtained by polymerase chain reaction (PCR) with the following primers, 5′-ATGGATTCTAGAACAGTTGGTAT-3′, and 5′-TTACTTGTTTTCCAGACAAGCTTC-3′. For nucleotide sequence analysis of the C. boidinii ADE2 gene, nested deletion mutants were prepared by a modified method of Yanisch-Perron (Sambrook et al., 1989). DNA sequence was determined with the automated DNA sequencing method involving PCR (373A DNA sequencer, Applied Biosystems) using fluoresceined primers. Transformation of C. boidinii and S. cerevisiae was performed by the lithium acetate method (Ito et al., 1983).
Construction of a plasmid for disruption of the ADE2 gene
A 2.6 kb SalI–PstI fragment containing the C. boidinii URA3 gene (Sakai et al., 1991 and1992b) was cloned between the SalI and PstI sites of pBluescript II SK− and the resultant plasmid was designated pCBU3. The 5′ untranslated region of the URA3 gene was isolated as a 0.9 kb SalI (blunt-ended)–XbaI fragment from pCBU3, and ligated between the KpnI and XbaI sites of pUC19 with the 2.6 kb KpnI–PstI (blunt-ended) fragment pCBU3. The resultant plasmid, pURP, contained a direct repeat of 0.9 kb fragment of the URA3 gene (Figure 1). XhoI linkers were inserted into the EcoRI site, localized in the coding region of the ADE2 gene of pCBAD2 (Figure 1), and the 3.5 kb of SalI fragment of pURP was ligated at this XhoI site. This resulted in plasmid pDAU, which was used for ADE2 gene disruption (Figure 1).
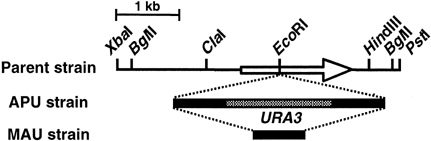
Restriction map of the cloned fragment containing the C. boidinii ADE2 gene and gene disruption of the C. boidinii ADE2 gene. Arrow indicates the position and orientation of the open reading frame
Results and discussion
Cloning of Candida boidinii ADE2 gene
In order to clone ADE2 gene from C. boidinii KST25 genomic DNA, Southern analysis was carried out using the radio-labelled S. cerevisiae ADE2 gene as a probe. Hybridization under conditions of low stringency (6×SSPE, 55°C) revealed that the C. boidinii ADE2 gene was located in a 4.5 kb XbaI–PstI fragment. Genomic DNA of C. boidinii was digested with XbaI and PstI, DNA fragments at 4.5 kb position were purified from agarose gel, ligated between XbaI and PstI sites of pBluescript II SK−, and transformed into E. coli DH5α. Colony hybridization using the same probe was performed and a positive clone was obtained. Plasmid DNA (pCBAD2) was isolated from the transformant, and standard restriction analysis was carried out (Figure 1). The 4.5 kb XbaI–PstI fragment from pCBAD2 was isolated after T4 DNA polymerase treatment, and introduced into the blunt-ended BamHI site of plasmid YEp24. The resultant plasmid, YEp-CBAD2, was used to transform S. cerevisiae ade2 strain YPH499. All Ura+ transformants showed the Ade+ phenotype, indicating that the cloned DNA fragment contains the C. boidinii ADE2 gene and that it can complement the S. cerevisiae ade2 mutation.
Sequencing analysis of C. boidinii ADE2 gene
The nucleotide sequence of 2.6 kb ClaI–HindIII region from pCBAD2 was determined. We found an ORF of 1719 nucleotides coding for a polypeptide of 573 residues (Figure 2). As shown in Figure 3, comparison of the deduced amino acid sequence showed high homology to AIRC from Pichia methanolica (Hiep et al., 1993), Schwannionmyces occidentalis (Gourdon et al., 1995), Candida albicans (Tsang et al., 1997), S. cerevisiae (Stotz and Linder, 1990), Candida glabrata (Hanic-Joyce and Joyce, 1998), and Schizosaccharomyces pombe (Szankasi et al., 1988); 72%, 65%, 65%, 61%, 57%, 52% identity, respectively. The results that the cloned XbaI–PstI fragment complemented the S. cerevisiae ade2 mutation and that ORF in the DNA fragment encodes a protein, that is highly homologous to the AIRC enzyme of other yeast species, clearly indicate that the cloned gene is the C. boidinii ADE2.

Nucleotide and deduced amino acid sequences of the C. boidinii ADE2 gene. The deduced amino acid sequence is given beneath the nucleotide sequence of the ADE2 coding region

Comparison of the amino acid sequences of AIRC proteins from C. boidinii (Cb), P. methanolica (Pm), S. occidentalis (Sc), C. albicans (Ca), S. cerevisiae (Sc), C. glabrata (Cg), and Sz. pombe (Sp). Conserved amino acid residues are marked with (*)
Disruption of C. boidinii ADE2 gene
To establish the transformation system of C. boidinii employing a useful selective marker ADE2 gene, the chromosomal ADE2 gene was disrupted by integrative transformation. The C. boidinii ADE2 disruption vector pDAU was constructed as described in Materials and methods. In pDAU, C. boidinii ADE2 gene was interrupted by the insertion of the C. boidinii URA3 gene. Repeated sequences were contained upstream and downstream of URA3 gene (black box in Figure 1), enabling later regeneration of uracil auxotrophy after ADE2 gene disruption.
C. boidinii KST2515, the ura3 derivative of strain KST25, was transformed with BglII-digested pDAU, and red Ura+ transformants were obtained. Red transformants showed adenine auxotrophy, indicating that the ADE2 gene was disrupted. The Ade− Ura+ strain (APU) was grown in YPD medium, then 5-fluoroorotic acid (5-FOA) resistant cells were selected (Alani et al., 1987; Sakai and Tani, 1992a). Strain MAU, which had been picked up as a 5-FOA resistant colony, showed the Ade− Ura− phenotype due to the loss of URA3 sequence. Strains APU and MAU were confirmed to have the correct gene replacement at the ADE2 locus by Southern analysis (data not shown).
The transformant in which the ADE2 gene was disrupted can be used as a host for transformation of C. boidinii, using the new ADE2 marker gene cloned in this study. The ADE2 gene can be used as a marker for transformation since it confers a white colour on transformants, and it allows the selection of Ade+ transformants among Ade− red-coloured colonies. For example, loss of the marker gene is easily detected by monitoring red segregants lacking ADE2 gene in a lawn of white colonies.
Acknowledgements
We thank Dr Keiji Kondo for helpful discussion and for help in revising the manuscript.




