Drawbacks of GENEHUNTER for larger pedigrees: Application to panic disorder
Abstract
Large pedigrees can pose a problem for GENEHUNTER linkage analysis software. Differences in two-point and multipoint lodscores were observed when comparing GENEHUNTER to other linkage software. Careful consideration must be given when selecting linkage analysis programs. Am. J. Med. Genet. (Neuropsychiatr. Genet.) 96:781–783, 2000. © 2000 Wiley-Liss, Inc.
INTRODUCTION
The linkage analysis community has become increasingly aware of some of the ways in which we lose linkage information when we discard or modify certain components of our data. Numerous studies have examined different aspects of this problem in recent years; one need only look at recent Genetic Analysis Workshops [e.g., Greenberg, 1999; Wijsman and Amos, 1997] to see some current examples. We became particularly interested in what happens when one uses GENEHUNTER to analyze pedigrees with complex structures that the computer program cannot handle exactly. The results of a multipoint linkage analysis of panic disorder [Goedken et al., 1999] using GENEHUNTER prompted us to examine the capabilities and limitations of the GENEHUNTER program. Here we show how trimming and/or splitting pedigrees can significantly reduce the amount of linkage information in the pedigree.
METHODS
In preparation for joint linkage analysis, we analyzed data from two respective genomic screens (University of Iowa and Columbia University) (Crowe et al., in review) [Knowles et al., 1998]. We calculated multipoint lodscores using GENEHUNTER v. 1.2 [Kruglyak et al., 1996; Kruglyak and Lander, 1998] for several markers. The GENEHUNTER output showed that due to time and memory constraints, several families were ‘trimmed’ (i.e., the program removes individuals in a pedigree until the pedigree is small enough to be analyzed). The documentation for GENEHUNTER warns that larger pedigrees will be trimmed unless they are skipped completely and recommends dividing the large pedigrees into two or more smaller pedigrees that can be analyzed in full. We were interested in the effects of trimming or splitting the larger pedigrees—in particular, the effects on the maximum model-based lodscore and the estimation of the recombination fraction. We explored these effects by analyzing the data using two methods. First, we calculated two-point lodscores using MLINK from the LINKAGE package [Lathrop et al., 1984] and GENEHUNTER and compared the results. We then calculated multipoint lodscores using VITESSE [O'Connell and Weeks, 1995] and GENEHUNTER and compared the results.
For the analyses, we selected six larger pedigrees from the original set of pedigrees. Four families showed evidence for linkage and the remaining two evidence against linkage. The pedigrees averaged 26 individuals with a range of 17–39 individuals, and between 3–13 affected individuals per family.
Additional settings can be specified when using the GENEHUNTER program. For example, we set the “skip large” option to “off,” thus allowing GENEHUNTER to trim the large pedigrees but still use them in the analysis. Another setting is the ‘max bits’ parameter, which must be set to keep computations within the ability of the computer. We set max bits to 14 because a larger value caused an error on our workstation. The max bits option refers to the formula 2N − F, where N = nonoriginals (individuals with parents in the pedigree) and F = founders (individuals with no parents in the pedigree). The “discard” option can also be set to on/off. We set it to off so that unaffected offspring of informative parents, that do not themselves have descendants, were not automatically discarded from the analysis.
RESULTS
- 1.
MLINK using the six whole pedigrees = −2.6.
- 2.
GENEHUNTER trimming the six pedigrees = −1.6.
- 3.
GENEHUNTER using six pedigrees we split into 20 = −1.2.
Splitting the pedigrees did not fare well and did not preserve pedigree information, so from here on we will focus our comparisons on the GENEHUNTER-trimmed pedigrees.
A substantial loss of information is evident when larger pedigrees are automatically trimmed during analysis. For instance, an MLINK two-point analysis on a pedigree with 39 individuals resulted in a lodscore of −2.0 at θ = 0, but the GENEHUNTER two-point analysis resulted in a lodscore of only −0.56 at θ = 0. In this particular pedigree, GENEHUNTER trims 17 individuals, including six affected individuals.
Another family, which includes 24 individuals, demonstrates two effects of GENEHUNTER trimming. During analysis, GENEHUNTER trims 11 individuals. None of the individuals trimmed are affected but two informative unaffected individuals are removed from the analysis. One consequence is a loss of information. The MLINK two-point lodscore at θ = 0 is +0.34, whereas the GENEHUNTER two-point lodscore at θ = 0 is only −0.04. A second consequence is an error in the maximum homogeneity lodscore. The MLINK maximum occurs at θ = 0, while the GENEHUNTER maximum occurs at θ = 0.5, producing a substantial error in the estimation of θ.
Two of the four remaining families exhibited lodscore differences when comparing GENEHUNTER to MLINK. In the first family, the MLINK lodscore at θ = 0 is −0.56, while the GENEHUNTER lodscore is −0.05. The MLINK lodscore for the second family is +0.74 at θ = 0, whereas the GENEHUNTER lodscore is +0.26. The last two families have similar lodscores across all values of θ when comparing GENEHUNTER to MLINK.
GENEHUNTER has the capability of using many markers in multipoint calculations, but at the expense of trimming data. Thus, we explored the effects of trimming by comparing GENEHUNTER multipoint lodscores to VITESSE multipoint lodscores. VITESSE can include all individuals from one set of founding parents in its calculations, thus using more of the pedigree information. VITESSE has a limitation on the number of markers, so four markers common to Columbia and Iowa were used for this comparison. GENEHUNTER multipoint lodscores were calculated using these four markers and all common markers on a particular chromosome with GENEHUNTER trimming the pedigrees.
The multipoint lodscore results spanning the four markers (17 cM) are shown in Figure 1 for one of the six families (Family X). The VITESSE lodscore calculation across the four markers (V-4) is more indicative of the information available in this particular region compared to the GENEHUNTER analyses using the four markers (GH-4) and all of the markers (GH-All). This demonstrates a drawback of using GENEHUNTER when the families collected are large, as a loss of information due to trimming the pedigrees can produce errors in the maximum lodscore. Furthermore, the GENEHUNTER analysis using all markers does not extract much more information than when four markers are used. Thus, in some situations it is beneficial to use fewer markers and all of the pedigree information.
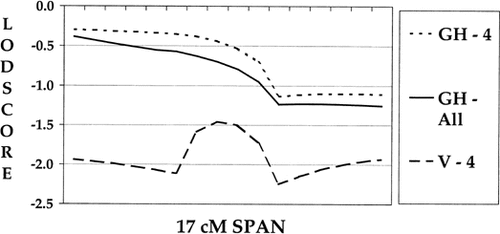
Multipoint results comparing GENEHUNTER and VITESSE results on one of the six families.
DISCUSSION
GENEHUNTER and VITESSE are two software programs that are frequently used for model-based multipoint analysis of extended pedigrees. Each program has advantages and disadvantages. The GENEHUNTER program is convenient to run and customize, many markers can be analyzed, and the computation time is reasonable. However, the advantages come at the expense of losing pedigree information since GENEHUNTER can only handle moderate-sized pedigrees. Moreover, the GENEHUNTER recommendation to split pedigrees can result in a greater loss of information than using the trimming method. Furthermore, splitting the pedigrees has a disadvantage of invalidating the heterogeneity lodscore. VITESSE calculates exact likelihood values and can handle larger pedigrees. However, VITESSE computations can be time-intensive, and the number of markers that can be used for analysis is limited.
In summary, the choice of statistical software for multipoint linkage analysis needs careful consideration. The capabilities and limitations of each software package need to be weighed against practical issues, such as type of data collected, the questions that need to be answered, and whether the software will run on the available equipment in a reasonable amount of time. We have shown here that the use of GENEHUNTER on larger pedigrees can lead to significant loss of linkage information and/or errors in the estimation of θ. We encourage more investigators to make comparisons of the available software to enable informed decisions about which program to select for any given situation.
Acknowledgements
We thank the two anonymous reviewers for their helpful suggestions.




