Identical distribution of the α2-macroglobulin pentanucleotide deletion in subjects with alzheimer disease and controls in a German population
Abstract
Recently, an association between a deletion polymorphism in the α2-macroglobulin gene (A2M) and Alzheimer disease (AD) has been reported. The aim of the present study was to corroborate this association in a German population of 102 AD patients and two control samples of 191 healthy subject and 160 depressed patients. The frequency of the A2M genotype in AD patients was almost identical to that in both control samples. Logistic regression analysis revealed an effect of age and the APOE genotype on AD risk, but no effect of the A2M genotype. Our findings do not support the fact that the previously reported positive association between A2M deletion polymorphism and AD modifies the disease risk in the studied population. Am. J. Med. Genet. (Neuropsychiatr. Genet.) 96:775–777, 2000. © 2000 Wiley-Liss, Inc.
Alzheimer disease (AD) is the most frequent cause of dementia in the elderly. The apolipoprotein E (APOE) ϵ4 allele is hitherto the only widely accepted major genetic risk factor associated with the late-onset familial and sporadic forms of AD. The gene (A2M) that encodes the proteinase inhibitor α2-macroglobulin (α2M) was considered a promising candidate associated with the risk for AD. Several findings suggest that α2M is pathophysiologically related to AD in many ways: 1) α2M is a constituent of the senile plaques and is upregulated in brains of AD patients or following brain injury [Bauer et al., 1991]; 2) α2M binds to beta-amyloid peptide (Aβ) and mediates the clearance of the α2M–Aβ complex via endocytosis [Du et al., 1997]; 3) α2M reduces the formation of Aβ fibrils by enhancing the solubility of Aβ [Du et al., 1998]; and 4) inflammatory cytokines implicated in AD stimulate the synthesis of α2M [Ganter et al., 1991].
Two variants of the α2M gene (A2M), an intronic pentanucleotide deletion and a G → A transition (Val1000Ile), were recently found to be associated with AD [Blacker et al., 1998; Liao et al., 1998]. The highest association with AD was found for the A2M deletion in a family-based association with 104 families by Blacker et al. [1998] (conditional odds ratio = 3.56). A marginally significant association was found when the same original sample was reanalyzed by another group [Rudrasingham et al., 1999] and in an independent sample [Dodel et al., 2000]. However, most case-control or family-based association studies failed to confirm the reported association between the A2M deletion polymorphism and AD [e.g., Dow et al., 1999; Rogaeva et al., 1999].
The divergent results in different populations underlines the importance of population admixture. Consequently, the aim of the present case-control study was to corroborate the earlier finding of an association between the A2M deletion and AD in an ethnically homogeneous German population living in the same region.
The study protocol was approved by the Ethics Committee of the Faculty of Medicine of the University of Bonn. All participants gave informed consent. Three independent groups of subjects were studied: AD patients, healthy subjects, and depressed patients. Population admixture was kept at a minimum by the recruitment of German patients and controls living in the same region. With the support of the local Census Bureau a stratified random sample of subjects over 50 years was selected from the general population. The depressed and AD patients were consecutively recruited from the Department of Psychiatry of the University of Bonn.
Probable AD was diagnosed according to the NINCDS-ADRDA criteria [McKhann et al., 1984]. Medical and family history, general medical and neurological examination, psychiatric interview, neuropsychological testing, blood and cerebral spinal fluid studies, and CT scans were carried out to exclude other forms of dementia. The AD and depressed patients underwent the same clinical evaluation. CT scans and cerebral spinal fluid studies could not be performed in the control group of healthy subjects. In this group, dementia was excluded by psychiatric interview and neuropsychological testing.
The mean age of the 102 AD patients was 74.4 ± 10.3 years (range, 51–101 years); 67% were female. The control group of 191 healthy subjects (52% females) had a mean age of 70.6 ± 11.4 years (range, 50–100 years) and the control group of 160 depressed patients (66% females) had a mean age of 68.0 ± 7.7 years (range, 50–88 years).
Genomic DNA was isolated from blood with the Quiagen blood isolation kit according to instructions provided by the manufacturer (Quiagen, Hilden, Germany). The A2M deletion and the APOE polymorphisms were detected using polymerase chain reaction (PCR) as described by Matthijs and Marynen [1991] and Hixson and Vernier [1990].
The A2M genotype and the allelic distribution were compared between AD patients and controls using χ2 test. Since AD patients were slightly older than controls, forward and backward logistic regression was performed for the simultaneous assessment of the effects of age (1-year intervals), gender (male or female), APOE genotype (APOE ϵ4 allele present or absent), and A2M genotype (A2M deletion allele present or absent), as well as interactions of these variables on AD risk. Statistical significance was established at P ≤ 0.05.
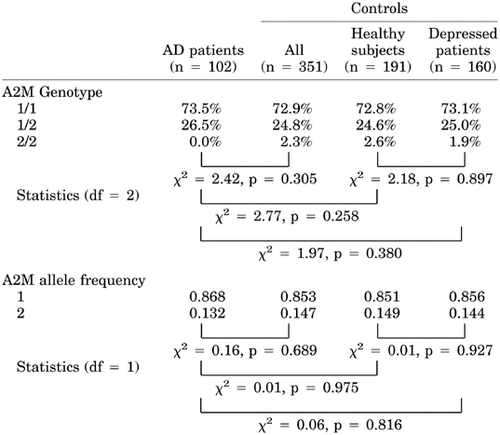
We could not find any significant difference in the A2M genotype or in the allele frequency between AD patients and all controls (genotype, χ2 = 2.42, df = 2, P = 0.305; allele frequency, χ2 = 0.16, df = 1, P = 0.689; Table I). This applied also for paired comparison between AD patients and healthy subjects, AD patients and depressed patients, as well as healthy subjects and depressed patients (P > 0.3). The distribution of the A2M genotype and the allele frequency in AD patients were almost identical to that of controls even after stratification of the groups into either APOE ϵ4 carriers (genotype, χ2 = 2.61, P = 0.279; allele frequency, χ2 = 0.25, P = 0.618), or noncarriers (genotype, χ2 = 1.13, P = 0.572; allele frequency, χ2 = 0.02, P = 0.904). Logistic regression revealed an independent effect of APOE genotype (odds ratio = 4.2, 95% CI = 2.5–7.1, P < 0.0001) and age (odds ratio = 1.05, 95% CI = 1.02–1.108, P = 0.0002) on AD risk, but no effect of the A2M genotype (P = 0.970).
| AD patients (n = 102) | Controls | |||
|---|---|---|---|---|
| All (n = 351) | Healthy subjects (n = 191) | Depressed patients (n = 160) | ||
|
|
||||
- a Allele 1 represents the wild type and allele 2 the pentanucleotide deletion.
Considering an effect size of 0.27, which is based on the frequencies of the A2M deletion allele previously reported for AD patients and unaffected individuals, the present study had a power of 80% (α-error = 0.05). We were not able to detect even a statistical trend; the distribution of A2M deletion allele was identical between AD patients and both control samples. Our results are in line with other studies that failed to demonstrate an association between the A2M deletion and AD [e.g., Dow et al., 1999]. Blacker et al. [1998] rebut that the discrepancies may be due to methodological differences, since in their study a statistical difference was observed only after applying family-based association methods. However, association studies in independent samples or samples that comprised some of the individuals also examined by Blacker et al. [1998] using the same statistical method (family-based association by the sibship disequilibrium test) also yielded divergent results [Rogaeva et al., 1999; Rudrasingham et al., 1999].
The biological relevance of the deletion is unknown. If the A2M deletion were a causal genetic risk factor for AD, replications of the observed positive association would be likely. However, it is possible that the positive association between the A2M deletion polymorphism and AD is due to a linkage disequilibrium with a nearby functional gene locus. In this case, divergent results in studies on different populations can be expected (as is the case in the present study), since linkage disequilibrium varies between populations.
In conclusion, our findings do not support the fact that the previously reported positive association between A2M deletion polymorphism and AD modifies the disease risk in the studied population.