Severe phenotypes associated with inactive ring X chromosomes
Abstract
Mental retardation and congenital malformations in individuals with small ring X chromosomes are often due to the functional disomy that results from failure of these chromosomes to undergo X inactivation. Such chromosomes either lack the XIST locus or do not express it. We have carried out genetic analysis of the ring X chromosomes from two girls with a 45,X/46,X,r(X) karyotype, mental retardation, and a constellation of abnormalities characteristic of the severe phenotype due to X disomy. In each case the ring X chromosome included an intact XIST locus which was expressed; the breakpoints were distal to DXS128, and therefore outside the XIC region; transcription analysis of alleles at the androgen receptor locus confirmed that these were inactive chromosomes. The characteristics of the XIST RNA were similar to the wild-type. Additional studies in cultured fibroblasts showed a second ring in a small percentage of the cells. The association of severe phenotype with an inactive X chromosome most likely reflects the presence of a second ring X chromosome which was active at least in some tissues during embryogenesis, but is no longer prominent in the tissues we analyzed. Am. J. Med. Genet. 93:52–57, 2000.
INTRODUCTION
Ullrich-Turner syndrome (TS) is characterized by short stature, gonadal dysgenesis, and a variable number of somatic abnormalities including webbing of the neck and lymphedema. Most often, mental retardation is not a manifestation of the syndrome [Van Dyke et al., 1991]. Approximately 50% of females with TS have a monosomy for the X chromosome with a 45,X karyotype. In the others there is a structural defect of one of the X chromosomes (46,X, delX); such individuals are often mosaic, having cells with 45,X as well. In some of the mosaic individuals, the deleted chromosome is a small ring X chromosome 45,X/46,X,r(X), and they usually have the classic TS phenotype [Zenger-Hain et al., 1993], as their ring X chromosomes undergo X inactivation, and are the inactive X in each cell. However, others exhibit intellectual impairment, growth retardation, and a series of congenital abnormalities, more severe than that usually seen with TS [Kushnick et al., 1987; Van Dyke et al., 1992; Dennis et al., 1993]. These include facial anomalies (hypertelorism, epicanthic folds, nystagmus, slanting palpebral fissures, broad nose, anteverted nares, dysplastic teeth), microcephaly, malformed lowset ears, chronic otitis media, soft tissue syndactyly, pigmentary dysplasia, hypotonia, scoliosis, seizures, EEG abnormalities, and malformations of the lung, kidney, and heart. The more severe phenotype in such cases can be explained by the inability of these small ring X chromosomes to undergo X inactivation, resulting in two active X chromosomes in cells with the ring X. Failure of X inactivation has been observed to result from deletion of the X inactive specific transcript (XIST)) gene [Migeon et al., 1993, 1994; Wolff et al., 1994; Jani et al., 1995] or deficient transcription of XIST [Migeon et al., 1993]. Further, Migeon et al. [1994] demonstrated that such chromosomes do have a functional disomy for genes present on the ring.
Yet some of the ring X chromosomes ascertained because of severe phenotypes do undergo X inactivation. Such inactive ring chromosomes are sometimes found along with an active ring X chromosome [Migeon et al., 1993; Jani et al., 1995], the duplication resulting from breakage and abnormal segregation of the rings during mitosis. We now report studies of two girls, showing that the single ring X chromosome present in most of their cells was the inactive X chromosome in these cells. The presence of a second ring X-chromosome in a few cells of both females, and the fact that ring chromosomes tend to be lost (as well as gained) during mitosis, suggest that the severe phenotypic findings we observed could result from functional disomy at the time when the phenotype was being determined in the embryo.
MATERIALS AND METHODS
Subjects
The two unrelated patients, SD and JD, were ascertained because their severe mental retardation and congenital abnormalities suggested the need for chromosome studies.
SD is the first child of healthy parents whose grandmothers are sisters. She weighed 2,085 g (<3rd centile) at birth, after 39 weeks gestation. Her psychomotor development was delayed from birth and she had recurrent upper and lower respiratory tract infections. Examination at age 11 months showed that she had sparse hair, abnormally modeled ears, protruding right ear and epicanthus, and an OFC of 44.2 cm (10th centile) (Fig. 1). She also had broad alveolar ridges, a left simian crease, and a right Sidney palmar crease. At age 2.5 years, she was hypotonic, unable to walk, and had no expressive language. There were no apparent abnormalities of her urinary amino acids, renal, cardiac, and neurological functions. Ophthalmologic examination showed strabismus convergens of the left eye and a CT-scan of her brain showed an abnormality in the occipital region with possible abnormal gyration. Radiographs of the spine showed thoraco-lumbar scoliosis. At age 4 she could walk with support and was on prophylactic antibiotics because of recurrent respiratory infections. There was no evidence of neck webbing or history of edema.
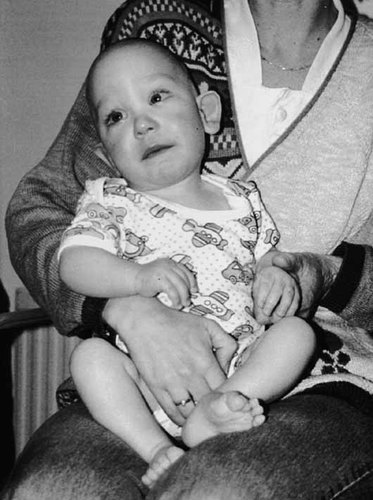
Photograph of SD at age 11 months showing protruding ears, strabismus convergens, long philtrum, and bilateral epicanthus.
JD was hospitalized at age 5 months for failure to thrive. During her first year of life she developed seizures which have continued with a frequency of 1–2 seizures/week. When evaluated for developmental delay at age 1.5 years, she had dolichocephaly, epicanthal folds, and a high arched palate. She later developed stereotypic hand movements and episodic hyperventilation. When seen at age 16, she had full-scale, verbal, and performance IQs of 45, 46, and 49, respectively, and was receiving special education. Menarche had not occurred and there were no signs of secondary sexual development. She was short of stature (136 cm, <5%) with prominent scoliosis. Her face was long and thin with a pouting mouth, long philtrum, down-slanting palpebral fissures, dacryocystitis, and high arched palate. Her hands and fingers were long and thin. She had decreased muscle tone but normal muscle strength and mass and brisk reflexes. She had a wide based gait with inturning of the left leg. She spoke in short sentences, with perseveration. Her hairline was normal with no evidence of neck webbing or cardiac abnormalities.
Cells and Clones
Primary human fibroblast cultures were established from skin samples of SD and JD and clones were obtained from the fibroblasts in early culture to enrich for the presence of the ring X chromosome as previously described [Migeon et al., 1985].
DNA Samples
For genotyping, DNA was prepared from blood samples from Patients 1 and 2, and from both parents of SD and the mother of JD.
Cytogenetic Characterization of Cell Lines
Peripheral lymphocytes or skin fibroblasts in their second or third passage were used for cytogenetic analysis. Metaphase spreads stained with GTG were used for chromosome analysis.
FISH Studies
To determine if the XIST locus was present on both X chromosomes, fluorescence in situ hybridization (FISH) was performed, using the cosmid XIST probe pXIST (ICRFc100H0130) which contains most of gene including the 3′ end and exon 6 [Lafreniere et al., 1993]. Further genotyping was carried out by FISH analysis with probes distal to XIST, which include P10Eg (ICRFc100F0138) and DXS128 (ICRFc100A1052). In addition, probes for the X centromere XCen [Walker et al., 1991] and pBAM-Xn were used to look for additional ring X chromosomes in interphase cells in Baltimore and Utrecht, respectively. Approximately 200 ng of each biotinylated probe was combined with 10 μg-cot-1 DNA. The probes were denatured at 80°C for 5 min. The metaphases were denatured with 70% formamide/2xSSC at the same temperature and time and slides were dehydrated in a series of cold ethanol rinses. Slides were hybridized overnight at 3°C, and washed with 50% formamide/2xSSC at 42°C for 5 min and 2xSSC at the same temperature. The signals were detected with three successive incubations with avidin and anti-avidin and chromosomes were counterstained with propidium iodide.
Ten to 20 metaphases with ring X chromosomes were analyzed for the presence of a signal on the ring chromosome. The signal from the normal X was used as an internal control.
RNA FISH using the cosmid XIST probe was carried out as described previously [Migeon et al., 1999].
Microdissection
In Case 1, five copies of the ring X chromosome were microdissected and amplified and used for FISH as described previously [Engelen et al., 1998]. For FISH studies the amplified microdissected material was fluorescently labeled by nick translation with biotin-14-dATP and this probe was used as a chromosomal paint. The results are shown in Figure 2.
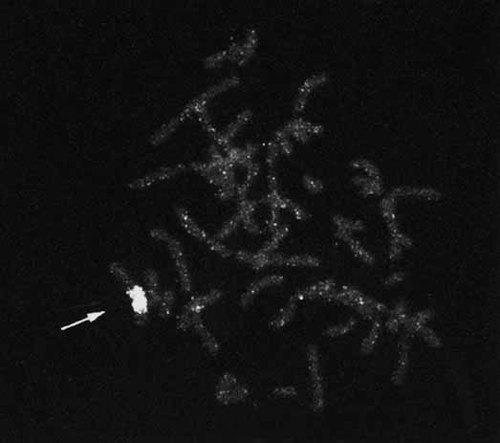
FISH painted metaphase from a male cell, showing content of ring X in SD. The probe was the DNA generated after microdissection and amplification of the ring chromosome, labeled with biotin-14-dATP. Note labeled material on both short and long arms of the single X chromosome (arrow).
Analysis of XIST Expression
RNA was isolated from cultured cells using TRIZOL reagent (Gibco/BRL, Gaithersburg, MD). cDNA was made from RNA using MuLV reverse-transcriptase (Perkin Elmer, Norwalk, CT) and oligo d(T) primers. PCR was performed with primers for XIST exon 6 and actin as an internal control as described previously [Migeon et al., 1994]. RNA samples were assayed in the presence and absence of reverse transcriptase. In addition, RNA FISH was used to examine the nature of the XIST transcript in fibroblasts from SD [Migeon et al., 1999].
Analysis of Transcripts at the Androgen Receptor Locus
To determine parent of origin and activity status of the ring X chromosome, we carried out transcriptional analysis using RT-PCR analysis of the polymorphic alleles at the AR locus [Allen et al., 1992] as described previously [Migeon et al., 1994].
RESULTS
Case 1 (SD)
Microdissection combined with FISH analysis indicated that the ring contained an X-chromosomal fragment consisting of parts of both short and long arms (estimated to include Xp21-cen-Xq21) based on reverse painting to metaphases from a normal male (Fig. 2). The prevalence of the ring X chromosome was determined by FISH using an X centromere probe; it was present in 22 of 43 metaphase cells and 300 of 800 interphase cells from the PHA-stimulated lymphocytes (46,X,r(X)), the others being 45,X. The ring was infrequent in the skin fibroblast cultures; of the 75 metaphases analyzed from the first biopsy, four had two ring chromosomes and one had 48 chromosomes, including three ring chromosomes (Fig. 3). The ring X was more prevalent in the second skin biopsy: 38 of the 136 interphase cells analyzed in Baltimore, and 25 of 100 analyzed in Utrecht. Two cells of the former and five cells of the latter included additional signals, possibly reflecting additional ring X chromosomes. With probes for the XIST locus, we observed signals on the ring as well as the normal X chromosome (not shown), with none of the 100 cells analyzed having more than two XIST signals (83 cells with one signal, 17 cells with two signals. A single ring X chromosome was present in three of 11 clones obtained from the fibroblast culture. FISH analysis with probes distal to XIST indicate that the p10EG and DXS128 loci are consistently present on the chromosome (Fig. 4A), placing the long arm breakpoint distal to DXS128, in Xq13.2, at least 200 kb telomeric to XIST [Lafreniere et al., 1993].
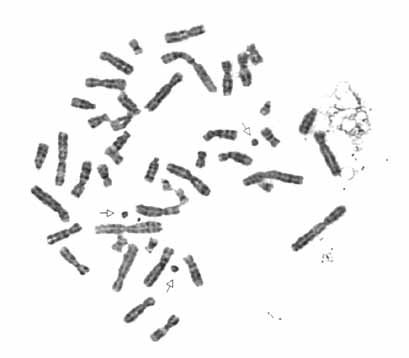
G-banded metaphase, containing three ring chromosomes (arrows) from SD fibroblasts.
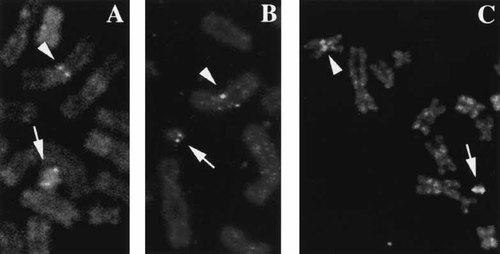
Ring X chromosomes from SD and JD include loci distal to XIST. Photomicrographs of FISH with probes for DXS128 (A, B) and p10EG (C), showing signals on rings from SD (A) and JD (B, C). Signals on rings are indicated by arrows, those on the normal X by arrowheads.
The results of RT-PCR analysis indicate that XIST is expressed in these cells (Fig. 5, top, lanes 6–9), presumably from the ring X chromosome. Studies of her alleles at the AR locus (Fig. 6, lanes 1–5) clearly show that SD inherited the ring allele from her father, as the lesser intensity (Fig. 6, lane 3, arrow) is consistent with its absence in the 45,X cells (38% in the SD specimen analyzed). Moreover, the RT-PCR analysis indicates only the mothers allele is expressed (Fig. 6, lane 4), as expected if the ring were an inactive chromosome.
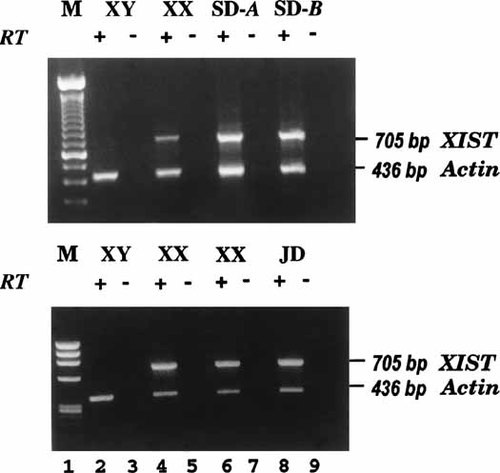
RT-PCR analysis showing that XIST is expressed in cells with the ring X chromosome. Skin fibroblasts from SD (top) and JD (bottom) amplified with primers for XIST exon 6, and actin as a control, in the presence (+RT) and the absence (-RT) of reverse transcriptase. Specimens: markers. (M) (lane 1), normal male, (XY) (lane 2–3); normal female, (XX) (lanes 4–5 and 6–7, bottom); SD, (two independent cultures) (lanes 6–9, top); JD, (lanes 8–9, bottom).
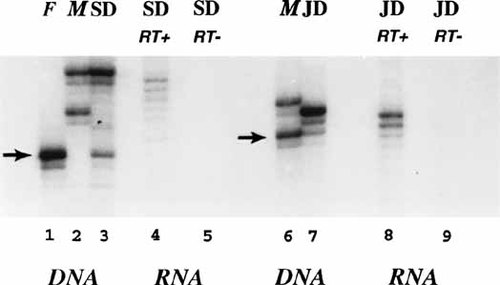
PCR analysis of alleles at the AR locus, to assay for parental origin and activity status of the ring X chromosomes. Lanes 1–3: blood DNA from SD (lane 3) her father (F) (lane 1) and mother (M) (lane 2). Lanes 4–5: RT-PCR using RNA from SD fibroblasts with (lane 4), and without (lane 5) reverse transcriptase (RT). Lanes 6–7: blood DNA from JD (lane 6) and her mother (M) (lane 7). Lanes 8–9: RT-PCR using RNA from JD fibroblasts with (lane 8) and without (lane 9) RT.
To determine if the XIST RNA was like the wild-type, we carried out RNA FISH on interphase cells from SD fibroblasts with the XIST probe. We found that the RNA was focused and stable to treatment with actinomycin D, behaving like XIST RNA from normal X chromosomes (Fig. 7).
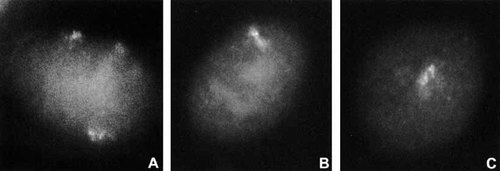
RNA FISH showing XIST RNA in interphase cells from SD. A: 49,XXXXY cell with three XIST RNA signals. B: SD cell with single XIST RNA signal. C: Another SD cell after 1-hr treatment with actinomycin D. Note: signals from the SD's ring X chromosome are similar in size and bipartite nature to those in the control cell and persist after actinomycin D.
Case 2 (JD)
In the case of JD, the small ring X chromosome (shown in Fig. 4B,C) was present in 50% of peripheral blood cells, 5% of cultured lymphoblasts, and in 80–43% of skin fibroblasts, the lower figure reflecting the decrease in ring frequency with time in culture. The ring X chromosome consistently labeled with the XIST cosmid FISH probe. Thirteen of 20 fibroblast clones obtained from her skin fibroblasts had the ring X chromosome in most cells (the ring is lost in some cells during propagation of the clone); some clones had cells with two ring X chromosomes, both containing the XIST locus. Both clones analyzed for XIST expression transcribed XIST (e.g., Fig. 5, bottom, lanes 8–9 ) Hybridization to probes for p10EG and DXS128, loci (Fig. 4B,C), indicate that the Xq breakpoint of the ring was distal to DXS128, which lies beyond the region shown to be sufficient for XIC function [Migeon et al., 1999].
Transcriptional studies of the polymorphic androgen receptor (AR) locus showed that the ring originated from her mother, as the paternal allele was predominant (Fig. 6, lane 7). The amount of the ring X allele in the DNA samples is consistent with its lower frequency in the specimen. With regard to transcription, clearly the AR allele from the normal X (the paternal one) is expressed in most cells (Fig. 6, lane 8). There are faint bands consistent with expression from the ring (maternal) allele (Fig. 6, lane 6, arrow); These faint bands are probably ‘shadow bands,‘ but they are compatible with the less likely possibility that the ring X allele is transcribed in a small number of cells. In any event, the faint bands are not consistent with the level of expression expected, as the preeminent ring was present in 80% of the cells in the specimen used as the source of cDNA.
DISCUSSION
Characteristics of the Ring X Chromosomes
We studied the ring X chromosomes from two girls ascertained because of their severe phenotype. Our attempts to find a population of cells with an active ring X chromosome were not successful, as the overwhelmingly predominant population of rings we studied in fibroblasts and blood cells had the characteristics of inactive X chromosomes.
Evidence That the Ring X Chromosomes Are Inactive
In both girls, the ring chromosome that could be analyzed is an inactive X chromosome, based on the fact that it expressed XIST, and did not express the AR locus on the ring X chromosome. We also show that the XIST RNA transcribed from the ring X chromosome is focalized around the chromosome as expected for an inactive X.
How Does One Reconcile Their Phenotype With the Inactivity of the Chromosome?
Unexplained mental retardation is seen in some individuals with TS, in association with a variety of karyotypes [Van Dyke et al., 1991; Sybert, 1995] including inactive ring X chromosomes [Yorifuji et al., 1998]. However, the abnormal phenotype found in our patients is more severe than that observed in TS [Van Dyke et al., 1991] and is reminiscent of the phenotype due to transcriptionally active ring X chromosomes. The constellation of abnormalities seen with functional X chromosome disomy varies considerably, because of the different genetic content of the rings [Jani et al., 1995], but there are some common traits. Many of the abnormal findings in our patients have been observed in individuals with active rings. On the one hand, this could be merely fortuitous. On the other, it may signify the presence of an active ring during the time in embryogenesis when these phenotypic features were being determined. Conceivably, a deleted form of the ring X chromosome, missing sequences essential for X inactivation, was produced prior to X inactivation during replication of the ring in somatic cells. This chromosome would be at a selective disadvantage because of its inability to undergo X inactivation. Cells which eliminate the chromosome by mitotic errors, such as breakage or nondisjunctional events, would be favored and give rise to a population of 45,X cells. Even the inactive ring X chromosome in these two patients tended to be lost during propagation of their cells in culture. Based on 1) their phenotypic resemblance to females who have active ring X chromosomes, 2) the presence of more than one ring in some of their cells, and 3) the fact that ring chromosomes are often lost during cell division, it seems likely that the severe phenotype in these females reflects the presence of a significant number of active ring X chromosomes in some tissues at the critical time during embryonic development.
What Is the Significance of the Findings With Regard to Prenatal Detection of Ring X Chromosomes?
Detection of ring X chromosomes during prenatal testing for genetic abnormalities raises the question of their significance with respect to clinical abnormalities. Will the phenotype be TS only, or will it be more severe? The results of our studies suggest that it may not be possible to accurately predict prenatally the phenotype that will be associated with the ring X chromosome after birth. Whereas the demonstration that the ring X is an active X chromosome (XIST is not expressed) means that the fetus at risk is likely to have severe phenotypic abnormalities, the demonstration that the ring X chromosome is an inactive chromosome (XIST is expressed), is not necessarily reassuring. One must consider the possibility that an active ring X chromosome was present early in embryogenesis, but is no longer present. As a corollary, correlations between the genotypes of rings found after birth and clinical phenotypes may be misleading,
Acknowledgements
We thank Joyce Axelman and Ashis Chowdhury for their contributions to these studies. We also thank MLW van der Heiden for referring patient SD. This work was supported by NIH grant HD05465 from NICHD to BRM




