Molecular cloning and characterization of a glucan synthase gene from the human pathogenic fungus Paracoccidioides brasiliensis
Abstract
1,3-β-D-glucan is a fungal cell wall polymer synthesized by the multi-subunit enzyme 1,3-β-D-glucan synthase. A subunit of this integral membrane protein was first described as the product of the FKS1 gene from Saccharomyces cerevisiae using echinocandin mutants. Other FKS1 genes were also reported for Candida albicans, Aspergillus nidulans and Cryptococcus neoformans. Here, we report the nucleotide sequence of the first homologous FKS gene cloned from the pathogenic fungus Paracoccidioides brasiliensis. An open reading frame of 5942 bp was identified in the complete sequence, interrupted by two putative introns, the first close to the 5′ end and the second close to the 3′ end of the gene. A promoter region is also described containing consensus sequences such as canonical TATA and CAAT boxes and, possibly, multiple sites for glucose regulation by creA protein. The deduced sequence of 1926 amino acid show more than 85% similarity to FksAp from A. nidulans, and 71% to Fks1p and Fks2p from S. cerevisiae. Computational analysis of P. brasiliensis Fks1p suggests a similar structure to transmembrane proteins, such as FksAp, with the presence of two domains composed by hydrophobic helices that limit the putative highly hydrophilic catalytic domain within the cytoplasm. The complete nucleotide sequence of PbFKS1 and its flanking regions have been submitted to GenBank under Accession No. AF148715. Copyright © 2000 John Wiley & Sons, Ltd.
INTRODUCTION
Paracoccidioides brasiliensis is a fungus of medical importance in rural areas of Latin America, causing a human mycosis (Lacaz et al., 1991; Brummer et al., 1993). The fungus shows a thermal dimorphism, changing from mycelial form (22°C) to yeast form (37°C), which is crucial for the establishment of infection in the human host (Franco, 1986; Lacaz et al., 1991). P. brasiliensis grows as a saprobic on soils, and the inhalation of its mycelium and/or its propagules is believed to be the most common infection pathway (Restrepo, 1985). Conversion to yeast cells occurs in the lungs, establishing the infection (Franco, 1986).
The fungal cell wall forms an interface between the parasite and the host. It has been suggested that this interaction may have an important role in virulence (San Blas and San Blas, 1985). The glucan component of the P. brasiliensis cell wall differs in the yeast and mycelial forms. It was observed that β-glucans are present in the mycelium while α-glucans are prevalent in the yeast cell wall (Kanetsuna et al., 1969), which indicates a possible role of cell wall enzymes in the fungal dimorphic transition. In this way, enzymes related to cell wall synthesis are of great interest in studies of dimorphism and its implications. In addition, new drugs such as those of the echinocandin family, have been described as potent inhibitors of 1,3-β-D-glucan synthases in several fungi (Debono and Gordee, 1994) which is highly significant in respect to paracoccidioidomycosis.
In this paper, we report the sequence of the first cloned P. brasiliensis 1,3-β-D-glucan synthase gene (PbFKS1). The presence of additional PbFKS genes is discussed.
MATERIALS AND METHODS
Media and strain
The clinical isolate of P. brasiliensis, Pb01, was kept at 37°C and subcultured in Fava-Neto medium (Fava-Neto, 1955).
Preparation of total DNA
After 7 days incubation at 37°C, yeast cells were scraped from slants, extensively washed with sterile water and frozen in liquid nitrogen. About 5 g of cells were disrupted by grinding to a fine powder. The total DNA was prepared according to Del Sal et al. (1989) with few modifications. Briefly, the cell powder was added of 10 ml extraction buffer (2% polyvinylpolypyrrolidone; 1.4 M NaCl, pH 8.0; 0.1 M Tris–HCl; 0.02 M EDTA; 2% CTAB). After incubation at 65°C for 1 h, three extractions with chloroform/isoamyl alcohol (v/v) were performed and nucleic acids then precipitated with ethanol. After RNAse treatment, the DNA was precipited with ethanol, centrifuged and resuspended in sterile water.
Homologous probing
PCR reactions were performed using oligonucleotides: E5, CA(G/A)ATGTT(C/T)GGTGGTAAT(T/A)CTG; E3, CATTTG(C/T)TCACCCAT ACCA(G/A)(T/C)GCC; I5, GCTTT(G/A)ATTGATGA TGG(A/T)CA(C/T)TGTG and I3, GAGAAAAT(G/A) TATTCAC(G/T)GGC(A/G)CC. The letter E refers to external primers and I to internal ones which permitted the nested PCR. The 25 µl buffered reaction mixtures contained 0.5 µM each primer, 1.5 mM MgCl2, 2.5 mM each dNTP, 2 U Taq DNA Polymerase (Cenbiot, Brazil) and 2.5 ng P. brasiliensis total DNA. Amplification conditions were: one cycle 94°C for 3 min, 54°C for 2 min and 72°C for 1 min and 30 s. 35 cycles were performed at 94°C, 1 min and 30 s, 54°C 1 min and 30 s and 72°C, 1 min and 30 s. The final extension was done at 72°C for 10 min. For nested PCR using the internal primers, 1 µl of the previous reaction was used as template. The amplified fragment was isolated with the Glassmax™ Kit (Gibco-BRL Inc., USA), subcloned in pGEMT-Easy vector (Promega, Co., USA) and sequenced using the T7 sequencing kit (Amersham Pharmacia Biotech, USA).
Construction of a genomic library
P. brasiliensis total DNA (10 µg) was partially digested with Sau3AI in order to generate fragments around 17 kb in length. The fragments were fractionated on 1% agarose gels and purified by electro-elution before cloning into the arms of λ DashII vector (Stratagene). The recombinant phage were cultivated according to the manufacturer's protocols. Around 3.5×108 pfu/ml were obtained and a library amplification resulted in a titre of 1012 pfu/ml.
Isolation, subcloning and sequencing of putative FKS sequences
Screening of the amplified genomic library was performed using a homologous probe in the form of a 300 bp PCR fragment isolated previously and labelled using the random prime labelling kit (Amersham Pharmacia Biotech, USA) and [α32-P]dCTP. The DNA from the selected clones was obtained as described by Grossberger (1987) and digested with restriction enzymes. Three different profiles of digestion were obtained. One clone of each profile was chosen to be analysed by Southern blot. The clones were named C1, C2 and C3. Hybridization was carried out in 45% (v/v) formamide, according to Sambrook et al. (1989). The washing stringency was 2×SSC, 0.1% SDS at 37°C. The DNA fragments were subcloned into pUC18 and sequenced. Partial sequences were first obtained by the manual procedure in order to define the clones of interest. Further analysis were performed by automated sequencing by primer walking (Core Facility for Protein/DNA Chemistry, Queen's University, Kingston, Canada).
Nucleotide sequence analysis
The fragments of clones C1, C2 and C3 that hybridized to the Southern blot were highly homologous. Total assembly of the PbFKS1 sequence was done using a sub-library of the EcoRI fragments of clone C2 digestion. Oligonucleotides EC3, TGTCAGAATCGCGTTGAACC and EC4, GCCATCGAAAATTTCATATCCC were designed for the amplification of the PbFKS1 sequence (same conditions as described above) spanning the region comprised by the internal EcoRI. The fragment was cloned into pUC18 vector and sequenced.
DNA and deduced amino acid sequences were analysed using the Genetics Computer Group (GCG) package (Madison, WI, USA). The topology of the transmembrane Pbfks1 protein was analysed according to von Heijne (1992), using the Top Pred2 programme available at the GCG package.
RESULTS AND DISCUSSION
Isolation and determination of the P. brasiliensis FKS1 nucleotide sequence
In order to obtain a P. brasiliensis genomic fragment coding for a conserved region reported for FKS genes, four oligonucleotides were designed on the basis of the homology among the catalytic regions previously reported sequences (Douglas et al., 1994; Mazur et al., 1995; Kelly et al., 1996). Nested PCR was done with oligonucleotides I3 and I5 using the PCR reaction (E3/E5) as template. This resulted in a 300 bp product which was cloned and used to screen the genomic library of P. brasiliensis. Several positive clones were found, from which some were selected for DNA digestion. Three clones were subjected to single (EcoRI) and double (EcoRI/PstI) DNA digestions. The restriction analysis showed distinct patterns among the selected clones. This was followed by Southern blot analysis of these clones using the 300 bp PCR fragment as a probe. Three different patterns of hybridization with the homologous fragment (Figure 1A, lanes 1–3) were detected by EcoRI digestion of clones 1, 2 and 3. Double digestion EcoRI/PstI (Figure 1B), displayed just two distinct band sizes (lanes 1–3). The data suggest that the FKS-like sequences are located at two loci in the genome of P. brasiliensis.
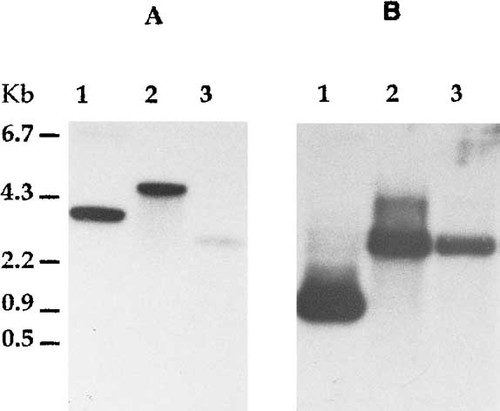
Southern blot analysis of selected clones from the genomic library. The DNA from the selected clones was digested with EcoRI (A) or EcoRI/PstI (B), fractionated on a 0.8% agarose gel and transfered to nitrocellulose membranes. The hybridization was performed with a 300 bp homologous fragment previously PCR-amplified from P. brasiliensis genomic DNA, labelled with [32P]-dCTP. The selected clones were numbered 1, 2 and 3 (above figures). Sizes from λ EcoRI/HindIII DNA fragments are shown.
The positive EcoRI DNA fragments from clones 1, 2 and 3 (Figure 1A) were subcloned and sequenced. All the obtained sequences were highly homologous to FKS genes (Douglas et al., 1994; Mazur et al., 1995; Kelly et al., 1996). To obtain the entire gene sequence for clone 2, all the generated DNA fragments from its EcoRI digestion (7.0 kb; 4.5 kb; 4.0 kb and 1.8 kb) were subcloned into pUC18 and sequenced. The final nucleotide sequence is shown in Figure 2.

Nucleotide and deduced amino acid sequences of P. brasiliensis FKS1. EcoRI fragments from clone 2 were subcloned into pUC18 and sequenced. A total of 7380 nucleotides were sequenced and the predicted 1926 amino acids residues were deduced. Lower case letters refer to non-coding regions. Putative conserved regions related to TATA, CAT and polyadenylation signals are underlined; CREA and MIG boxes are double underlined; CT-boxes are bold; pH-responsive regulator motif is boxed; and PyAAG motifs are dotted. cAMP-responsive elements are indicated by wavy underlining. STRE and HSF motifs are dot dashed.The stop codon and possible N-glycosylation sites are labelled with an asterisk. The two introns are delimited by GT/AG and the lariat consensus sequences are shown. The PbFks1p domains 1 and 2 are underlined. The PTS motif is double underlined.
Nucleotide sequence of P. brasiliensis FKS1
The entire nucleotide sequence of P. brasiliensis FKS1 gene was obtained. A map of the gene is shown in Figure 3A, together with a PCR analysis (Figure 3B) of the region defined by EC3 and EC4 primers from total DNA and from clone 2 DNA. One can see that the amplified DNA of both samples shows the same size. Sequencing of EC3/EC4 fragments (data not shown) matches exactly with the corresponding region of PbFKS1 gene which eliminates the possibility of missing EcoRI fragments in the assembling of clone 2 sequence.
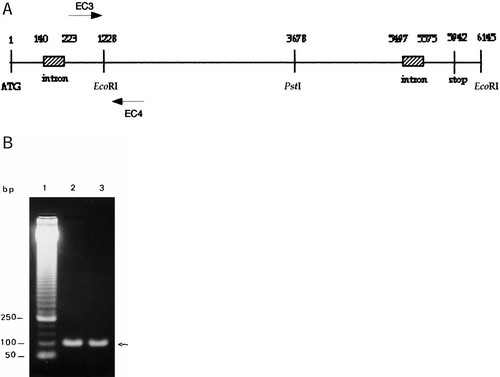
(A) Diagram representing the PbFKS1 gene structure. The localization of restriction sites and of oligonucleotides EC3 and EC4 are shown. (B) A 50 bp DNA ladder (lane 1) is shown. PCR gel analysis of total DNA (lane 2) and clone 2 (lane 3) with oligonucleotides EC3 and EC4. PCR conditions were: the 25 µL buffered reaction mixtures contained 0.5 µM of each primer, 1.5 mM MgCl2, 2.5 mM each dNTP, 2 U Taq DNA Polymerase and 2.5 ng P. brasiliensis total DNA or clone 2 DNA. Amplification conditions were: one cycle 94°C for 3 min, 50°C for 2 min and 72°C for 1 min and 30 s. Thirty-five cycles were performed at 94°C, 1 min and 30 s, 50°C 1 min and 30 s and 72°C, 1 min and 30 s. The final extension was done at 72°C for 10 min.
A putative promotor region of 1235 bp was analysed and presented several stop codons. The presence of sequences that are homologous to known regulatory elements from other micro-organisms in the PbFKS1 promoter region was detected. Nevertheless, no functional or regulatory significance of these elements has been demonstrated in P. brasiliensis.
Elements such as TATA (−126) and CAAT boxes (−244) are present. Two C/TAAG motifs are located above the putative ATG start codon (−52 and 121). These motifs are present in highly expressed S. cerevisiae genes near the mRNA transcriptional initiation site (Gurr et al., 1987). We also detected the presence of T-rich regions along the whole promoter sequence, mainly from nucleotides −562 to −935. Gurr et al. (1987) and Ballance (1986) described CT-rich regions as possible sites for RNA polymerase interaction in fungal promoter elements present in highly expressed genes. These CT boxes were localized in the PbFKS gene immediately downstream of the putative TATA box (−107).
At position −809 we found the sequence GCCAAG, which mediates pH-dependent gene expression in A. nidulans (Tilburn et al., 1995). This may be of importance during dimorphic transitions occurring during the establishment of host infection. The presence of these putative regulatory signals suggests a flexible and possibly complex control mechanism for the expression of P. brasiliensis genes related to cell wall assembly. CREB motifs (cAMP-responsive element binding protein) are also present (−54 and −1143). It has been suggested that cAMP may play a regulatory role in the first hours of mycelium-to-yeast transition in P. brasiliensis (Paris and Duran, 1985). Several HSF (heat shock factor) and STRE (stress-response element) motifs are found in the PbFKS1 gene. Two of them are shown in Figure 2 (−343 and −448). Several conserved regions related to MIG (−386) and CREA consensus (−432, −639 and −782) were also found to be similar to those of Aspergillus aculeatus (Takada et al., 1998) and A. nidulans (Cubero and Scazzocchio, 1994). Mazur et al. (1995) described the regulation of S. cerevisiae FKS1 and FKS2 genes by glucose. These authors found that the FKS1 transcript predominates during cellular growth on glucose, while FKS2 is expressed in the absence of glucose.
Three exons, separated by two putative introns, were detected. The first intron of 83 bp is located close to the 5′ end of PbFKS1, while the second one, of 78 bp, was found close to the 3′ end of the gene. All intervening sequences are flanked by 5′ GT and 3′AG, which correspond to the consensus sequence of known splicing sites. Both introns presented the splicing signal for lariat formation (Turner, 1993). The 3′ end shows a sequence rich in AT. An abbreviated form of the polyadenylation signal ATAA, present in filamentous fungal genes (Gurr et al., 1987), is also identified 154 nucleotides downstream of the TGA termination codon.
Deduced amino acid sequence
A deduced amino acid sequence of 1926 residues was found, starting in the first ATG in accordance to the Kozak rule (Kozak, 1981). A possible termination codon is located at position 5942. The complete open reading frame of 5781 nucleotides predicts a protein of 212 kDa. This molecular mass resembles the description of other Fks proteins, varying from 215 to 229 kDa (Douglas et al., 1994; Mazur et al., 1995; Kelly et al., 1996). Six potential N-glycosylation sites were found, two of which are located in the hypothetical catalytic region of the enzyme. Glycosylation is a very common post-translational modification in fungal enzymes and might confer stability to the protein, as is the case with the cellulases from Trichoderma reesei (Merivuori et al., 1985). These authors reported that N-asparagine-linked oligosaccharides do not appear to be necessary for its secretion or activity, but do seem to contribute to the stability of the enzyme.
Sites for the binding of glucose monomers were search in C. neoformans (Thompson et al., 1999), A. nidulans and S. cerevisiae (Kelly et al., 1996). No UDPG-binding motifs (QXXRW or R/KXGG) were verified. Similarly, this motif is absent in the putative catalytic region of PbFks1p. Kelly et al. (1996) described two transmembrane domains enclosing the catalytic region of FksA protein from A. nidulans by analogy to the cellulose synthase of Aerobacter xylinium. We also found similar domains (domains 1 and 2) in P. brasiliensis FKS1 gene, as indicated in Figure 2. Since cellulose synthase catalyses the formation of 1,4-β-glucan using UDPG as substrate, Kelly et al. (1996) suggest this region as a putative enviroment for UDPG binding without defining an specific site for UDPG linking.
The predicted PbFks1 protein also shows a PTS-HPr (phosphotransferase system–phosphoryl carrier protein) component phosphorylation site motif. The phosphoenolpyruvate-dependent sugar phosphotransferase system is a major carbohydrate transport system in bacteria (Meadow et al., 1990). PTS catalyses the phosphorylation of incoming sugar substrates, concomitant with their translocation across the cell membrane. In P. brasiliensis the synthesis and assembly of cell wall components seems to occur vectorally through the plasma membrane and into the periplasmic space, as discussed by Cabib et al. (1988). Thus, PbFks1p could be assembling the phosphorylated glucan polymer and simultaneously extruding it out of the membrane.
Comparison of the predicted PbFks1p to other Fksps
The deduced amino acid sequence of PbFks1p was compared with those described for S. cerevisiae and A. nidulans. The entire P. brasiliensis sequence showed 64% identity and 71% similarity with respect to the Fks1p and Fks2p of S. cerevisiae and 80% of identity and 85% of similarity with FksAp of A. nidulans. The catalytic domain showed a greater homology, as shown in Figure 4, reaching a value of 95% between PbFks1p and FksAp.
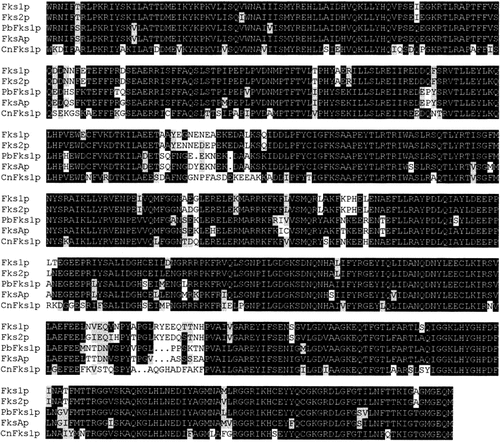
Multiple sequence alignment of the putative catalytic regions of Fks1p, Fks2p of S. cerevisiae (Accession No. U12893, U16783), PbFks1p of P. brasiliensis (Accession No. AF148715), FksAp of A. nidulans (Accession No. U51272) and CnFks1p of C. neoformans (Accession No. AF102882). Alignment of amino acids 796–1331 from the PbFks1p sequence corresponding to the proposed catalytic region was performed with the Pileup Program.
Hydropathy analysis by the Top Pred2 programme (von Heijne, 1992) predicted the PbFks1p as an integral membrane protein displaying about 16 transmembrane helices (HTM; Figure 5A). We found two putative helices (amino acid 646 and 1423). We consider the second in accord with the other Fksps described (Kelly et al., 1996). The N- and C-termini are both hydrophilic. The predicted catalytic domain of 588 amino acids residues is highly hydrophilic and a possible location in the cytoplasm is suggested. The catalytic domain is limited by two HTM domains with six and ten helices, the first at the N-terminal and the second at the C-terminal, as described for other Fksps (Kelly et al., 1996). The topology analysis (Figure 5B) was similar to other glucan synthases, with a large hydrophilic cytoplasmic domain of 588 amino acids. The number of amino acid residues found in Fks1p (582) and FksAp (578) cytoplasmic domains is also similar. The isoelectric point (pI) of the predicted PbFks1p is 8.5, which is in accordance to the pIs of the other homologues.
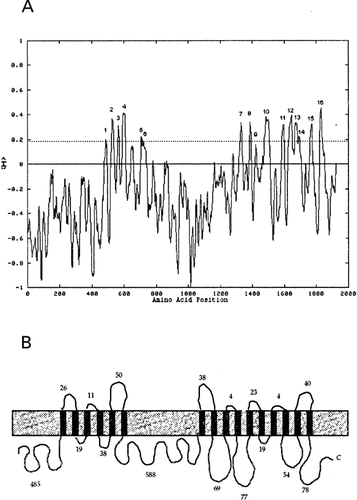
Hydropathy analysis of PbFks1p. (A) Hydropathy profile of PbFks1p by Top Pred2 programme (von Heijne) using a window of 21 amino acids and a core window of 11 amino acids. (B) Schematic topology diagram for the predicted transmembrane PbFks1 protein. The membrane is represented by a rectangle and the solid black line designates the polypeptide chain with its putative outer and inner loops. The 16 predicted transmembrane helices are indicate by vertical bars. The length (number of amino acids of the each non-transmembrane domain is shown. The grey line indicates hydropathy value (<H>) limit for transmembrane regions predicted. The numbers above each peak correspond to the length of the putative transmembrane domains.
In this paper we described the cloning and characterization of a P. brasiliensis glucan synthase gene, homologous to those described for S. cerevisiae (FKS1, FKS2 and FKS3) and A. nidulans (FKSA), among others (Thompson et al., 1999). Although we have fully characterized one gene, we have obtained two other DNA fragments which may correspond to possibly distinct genes (data not shown). In S. cerevisiae, three highly homologous FKS-like genes have been described, two of which perform distinct functions. The FKS1 gene is regulated in the cell cycle and predominates during growth on glucose. On the other hand, FKS2 is essential for sporulation and is expressed in the absence of glucose. In addition, this gene is induced by Ca, and is completely dependent on the Ca/calmodulin-dependent phosphoprotein phosphatase calcineurin (Mazur et al., 1995). The occurrence of more than one FKS-like sequence in P. brasiliensis may suggest a similar function for the FKS family in this parasite, such as differential gene regulation. We named our gene PbFKS1 on basis to the homology to the FKS family. However, the function of the P. brasiliensis gene characterized remains to be demonstrated. A better knowledge of this function will be of fundamental importance in targeting P. brasiliensis 1,3-β-glucan synthase genes by new drugs, such as those of the echinocandin family, described as efficiently antifungal in S. cerevisiae (Debono and Gordee, 1994). The promoter region analysis showed several putative motifs for creA protein binding, which is compatible with glucose regulation, as cited above. Preliminary RT–PCR experiments on PbFKS1 expression suggest that the transcript is not present in glucose-grown cells (data not shown), as is found for the FKS2 gene in S. cerevisiae (Mazur et al., 1995). Finally, further analysis of the complete promotor region of PbFKS1 will be necessary for a better understanding of its transcriptional regulation.
Acknowledgements
We thank Dr Peter Inglis for review and Maria de Fátima L. Cesário for laboratory assistance. This work was financed by CNPq/PADCT; PICD/CNPq and FUNAPE.




