Comparison of different microbial bioassays to assess metal-contaminated soils
Abstract
These experiments compared the sensitivity of four different types of bioassay over time after five metals were added to a wide range of soils at the maximum concentrations in the European Union Sewage Sludge Directive. Three were chronic assays (most probable number of Rhizobium leguminosarum, soil microbial C and Biolog substrate utilization). The fourth bioassay, an acute biosensor, employed a lux-marked luminescent bacterium (Escherichia coli) in the soil pore water. Five metals were added to 23 different soils as a mixture at Zn = 300, Cd = 3, Pb = 300, Cu = 135, and Ni = 75 mg/kg as nitrate salts and compared with unamended controls. Zinc and Cu were the metals most likely to be toxic at the concentrations used here. In the case of Rhizobium, the number of cells in soil was not affected after 11d; however, by 818 d the numbers had decreased by four orders of magnitude with increasing concentrations of Zn and Cu in soil solution. Microbial biomass also was not affected after 11d, but significantly decreased with increased Zn (p < 0.001) and Cu (p < 0.01) in soil solution after 818 d. Toxicity to the soil microbial biomass increased with time, whereas the toxicity to the biosensor remained the same. Biolog substrate utilization profiles were not responsive to the concentrations used here.
INTRODUCTION
Most current regulations governing metal toxicity in soils are based on the total metal concentration in soil. This approach fails to consider important soil properties that affect bioavailability of metals and, hence, is not relevant to risk or hazard assessment. New approaches are now required; for example, in the United Kingdom there has been a move toward risk-based assessment of pollution [1]. The pollutant source-pathway-receptor concept underpins the legislation behind this. In many cases a biological receptor is considered the important target, so methods are needed to assess impacts on those receptors rather than simply total metals in soil.
Assessment of contaminated sites is complicated by the fact that biological responses to the same concentrations of pollutants added to different soils vary according to the physico-chemical conditions, and many sites to be assessed are contaminated with more than one metal. The usefulness of various commonly used soil microbial bioassays across different soils that often are contaminated with mixtures of metals is not clear. In this paper we assess four bioassays and their suitability to be included in a suite of assays to assess the hazard of multiple metal contamination to microorganisms in soils. The inclusion of soil microorganisms in the hazard assessment process is important as this community governs the functionality of ecological processes in the soil environment. Disruption of this community may result in a degradation of soil health and the quality of the environment.
The assessment of the microbial community is complex, and as such must be carried out on several levels. The four bioassays assessed in this paper were chosen to represent different tiers of the microbial ecosystem. With this in mind, three solid phase bioassays were chosen to assess the chronic tox-icity of metals in soils. They were the quantification of the indigenous population size of the bacterium Rhizobium leg-uminosarum biovar trifolii, to assess the effect of metal contamination on an important plant symbiont, which supplies large amounts of fixed nitrogen in symbiosis with clovers; soil microbial biomass C, to assess the size of the whole microbial community; and substrate utilization, to assess the functional diversity of the bacterial community.
In addition, an aqueous-phase bioassay also was chosen to assess the acute toxicity of metals in the soil pore water. The bioassay used the luminescence-based bacterial biosensor Escherichia coli HB101 pUCD607.
Rhizobia are important in the supply of nitrogen to crops and the survival of an effective population has been shown by some authors to be sensitive to metals in soils. This was the case for R. leguminosarum bv. trifolii in the studies by Chaudri et al. [2, 3] and Giller et al. [4] and R. leguminosarum bv. viciae by Chaudri et al. [5]. The population of a specific biovar of Rhizobium in soil is measured using a bioassay with an appropriate test host species. Results, therefore, indicate the size of an individual population in soil.
All of the bacteria, fungi, and actinomycetes in soils have been referred to collectively as the soil microbial biomass [6]. Microbial biomass can be measured by techniques such as fumigation of the soil with chloroform, followed by extraction of the C or N that is released from the cells after fumigation. This measurement of the whole microbial community has been shown to decrease in response to metal contamination of soils [7, 8].
| Organic carbon (%) | Total Zn (mg/kg) | Total Cu (mg/kg) | Total Cd (mg/kg) | Total Pb (mg/kg) | Total Ni (mg/kg) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Soil series/location | pH (H2O) | ||||||||
| Land use | Soil type | ||||||||
| Bardsey | Deciduous woodland | Stagnogley | 3.3 | 3.2 | 29.0 | 5.2 | 0.03 | 83.3 | 6.7 |
| Wick | Deciduous woodland | Brown earth | 3.4 | 4.5 | 49.4 | 18.7 | 0.08 | 58.3 | 10.6 |
| Iveshead | Amenity heathland | Ranker/brown podzolic | 3.7 | 6.6 | 37.3 | 14.1 | 0.12 | 74.2 | 6.9 |
| Pwllpeiran | Agricultural, grassland | Clay loam | 5.2 | 3.0 | 206.0 | 14.3 | 0.28 | 74.0 | 24.9 |
| Braunschweig | Agricultural, arable | Silty loam | 5.5 | 2.3 | 76.7 | 15.7 | 0.26 | 27.6 | 13.1 |
| Fladbury | Agricultural, grassland | Pelo-vertic gley | 5.7 | 4.7 | 122.0 | 26.0 | 0.32 | 44.2 | 46.1 |
| Woburn | Agricultural, arable | Loamy sand | 6.3 | 1.0 | 39.3 | 8.4 | 0.1 | 27.5 | 12.5 |
| Ticknall | Agricultural, grassland | Cambic stagnogley | 6.3 | 5.2 | 172.0 | 19.7 | 0.55 | 375.8 | 21.7 |
| Cottam | Agricultural, arable | Stagnogley | 6.5 | 2.1 | 57.0 | 16.3 | 0.17 | 33.2 | 16.1 |
| Rosemaund | Agricultural, arable | Silty clay loam | 6.5 | 1.9 | 99.8 | 18.3 | 0.14 | 23.3 | 61.3 |
| Watlington | Agricultural, arable | Sandy loam | 6.6 | 1.2 | 44.0 | 9.4 | 0.14 | 24.3 | 19.9 |
| Rivington | Agricultural, grassland | Brown earth | 6.6 | 3.4 | 50.3 | 7.7 | 0.31 | 47.7 | 5.2 |
| Arrow | Agricultural, arable | Gleyic brown earth | 6.6 | 2.1 | 74.5 | 23.2 | 0.24 | 43.1 | 16.6 |
| Newport | Agricultural, arable | Brown sand | 6.6 | 1.0 | 45.9 | 8.0 | 0.16 | 33.3 | 9.3 |
| Insch | Agricultural, arable | Brown earth | 6.8 | 4.1 | 92.2 | 11.5 | 0.19 | 16.3 | 14.6 |
| Gleadthorpe | Agricultural, arable | Sandy loam | 6.8 | 1.7 | 53.3 | 10.4 | 0.12 | 22.6 | 6.4 |
| Denchworth | Agricultural, arable | Pelo-stagnogley | 7.0 | 4.4 | 62.7 | 25.0 | 0.27 | 38.1 | 34.7 |
| Marian | Amenity grassland | Humic rendzina | 7.2 | 7.1 | 139.0 | 38.7 | 0.3 | 83.6 | 23.0 |
| Worcester | Agricultural, arable | Argillic pelosol | 7.3 | 2.9 | 74.6 | 18.2 | 0.18 | 36.6 | 34.8 |
| Bridgets | Agricultural, arable | Silty clay loam | 7.3 | 2.4 | 94.3 | 11.1 | 0.98 | 34.0 | 40.1 |
| Evesham | Agricultural, arable | Calcareous pelosol | 7.3 | 2.4 | 66.4 | 22.3 | 0.54 | 36.5 | 30.5 |
| Hanslope | Amenity grassland | Calcareous pelosol | 7.5 | 4.4 | 102.0 | 18.6 | 0.24 | 33.7 | 29.1 |
| Ragdale | Amenity grassland | Pelo-stagnogley | 7.6 | 4.1 | 123.0 | 20.9 | 0.25 | 52.0 | 34.1 |
Biolog® profiling (Hayward, CA, USA) is a culture-based technique, and only relates to a limited fraction of the soil bacterial community [9]. The advantages are that it is relatively easy to perform and the results potentially are related to soil functional potential, although there is no direct relationship between C substrate utilization profiles and soil community functioning. Biolog was adapted for studying changes in the structure of microbial communities by Garland and Mills [10], including the effects of metals [11].
Constitutively marked bacterial biosensors have been developed that use bioluminescence as the transducer [12]. Bio-luminescence is a particularly useful reporter mechanism because it is sufficiently rapid to allow real-time monitoring, and it permits nondestructive, in situ measurement. Nonlumines-cent microbes can be marked constitutively with the lux-genes, allowing sensitive monitoring of cell metabolic activity and, hence, health. Two further benefits of bioluminescence make it suitable for environmental application. First, luminescence is rare in the terrestrial environment and, accordingly, interference is rare. Second, the measurement of luminescence reflects the metabolic activity of the cell.
Previous studies using these techniques usually have been based on different treatments of an individual soil or site [2-5, 7, 11, 13-16]. Our aim was to evaluate the effectiveness of these bioassays to assess the harm imposed by multiple metal toxicity of exactly the same total metal additions to a wide range of soils rather than in an individual soil. The aim, therefore, was not to investigate the toxicity of individual metals; in the data presentation, certain metals are selected on the basis of measurements of their solubility and previous information on their toxicity (where available). Bioassays were performed both soon after amendment and after two years of equilibration to give time for chemical stabilization to take place (aging) of the added metals, and to allow chronic effects on the microbial assays to be measured, more like the situation in the field. This could include recovery or increased toxicity.
MATERIALS AND METHODS
Microcosm set-up
Twenty-three uncontaminated soils (sampled from 0 to 15 cm) selected to cover a wide range of properties found in British soils were collected along with one German soil (from Braunschweig) (Table 1). A brief description is given below; a more detailed account has been given by Tye et al. [17].
A mass equivalent to 24 kg oven dry weight of each soil (moist, <4 mm) was divided between four 10-L plastic containers, which had pierced lids to allow gas exchange. For each soil, two control microcosms and two metal-amended microcosms were made. The metal-amended microcosms had five metals applied as nitrate salts, at concentrations representing the original ambient metal concentration in each soil plus an additional amount equivalent to the maximum concentration allowed by the European Union Sewage Sludge Directive [18] for soils in the pH range of 6 to 7 (Zn = 300, Cd = 3, Pb = 300, Cu = 135, and Ni = 75 mg/kg). The soils were then maintained at 80% field capacity at 16°C.
Sampling methods
Soil and soil pore water samples were taken from the microcosms 11, 67, and 818 d after the addition of metals. A representative soil sample was removed at day 11 and day 818.
After the soil was sampled for Rhizobium, biomass, and Biolog analysis, the moisture content of the soil was raised to 110% field capacity and maintained for 10 d. Soil pore water was then extracted using porous polymer Rhizon soil moisture samplers (Rhizosphere Research Products, Wageningen, Holland) under negative pressure (0.07 MPa) [19]. The microcosm soils were allowed to dry back down to 80% field capacity over approximately 10 d and rehomogenized; the incubation at 16°C then was continued until the next sampling time.
Microbial biomass C, substrate utilization, and rhizobial analyses were carried out at day 11 and 818 after metal amendment occurred. Rhizobia, Biolog, and biosensor analyses could not be carried out on the two to four most acid soils (Table 1). The reason for this likely is to be the small numbers of bacteria typically found in acid soils [20] and, in the case of the biosensor, luminescence is inhibited at low pH [21]. The most probable number (MPN) counts were not reproducible on the Rosemaund soil although the pH was 6.5, and no reason can be given for this. Biosensor analysis was carried out after 67- and 818-d incubation of the soils.
Chemical analysis of soil and pore water
Zinc, Cu, Ni, and Pb concentrations in the soil pore water were determined by inductively coupled plasma atomic emission spectrometry (Fisons-ARL Accuris, Ecublens, Switzerland) after acidification to 5% with HCl. Cadmium concentrations were determined by graphite furnace atomic absorption spectrometry (Perkin-Elmer ZL 4100, Norwalk, CT, USA) after acidification to 0.3% with HNO3. Metal concentrations in pore waters used for biosensor work were determined by inductively coupled plasma mass spectroscopy (ICP-MS, Var-ian Ultramass, Palo Alto, CA, USA) after the addition of 10 μl, 5 M HNO3 to 1 ml of sample and subsequent 10−1 dilution with deionized water. Correlation between the metal concentrations obtained by the different analytical methods was found to be very good (r2 = 0.99 for Zn, Cu, Ni, Pb, and Cd; all p < 0.001).
Soil pH was determined in deionized water (1:2.5 w/v). Soil organic C was determined by combustion analysis (LECO CNS 2000, LECO, St. Joseph, MI, USA).
Quantification of indigenous R. leguminosarum bv. trifolii
The MPN method was used to estimate the number of rhizobia in soil [3, 22] using Trifolium repens (white clover cul-tivar Grassland Huia) as a host. The MPN of rhizobia was calculated using MPNES computer program [23]. A decrease of more than one log unit was taken as significant [3].
Substrate utilization
The substrate utilization patterns of the soil microbial community were determined using Biolog™ Ecoplates. The plates contained 31 different C substrates (listed in [24]) and one blank well, all in triplicate. Preassessment of bacterial numbers in the soil samples taken at day 11 was carried out. Colony forming units were enumerated on nutrient agar plates inoculated with diluted soil suspension. The Biolog method was carried out as by Knight et al. [11]. Briefly, for soil extraction, the equivalent of 10 g oven dry weight of each moist soil was added to 90-ml sterile deionized water and shaken for 2 h at 200 oscillations per min. After standing for 3 min, each sample was diluted with sterile deionized water as necessary to give a final inoculum concentration in solution equivalent to 104 colony forming units/ml. Finally, a subsample of 140 μL of the diluted extract was added to each well with a multipipette. The plates were incubated at 28°C and read morning and evening using a Multiscan MCC plate reader (Labsystems, Helsinki, Finland). The time used for all subsequent data analyses was the first measurement after the start of exponential growth (measured by the average well color development). The soil solutions tested varied widely in pH, but it is known that color formation in the plates is not affected by the pH in different soil solutions [25].
Microbial biomass carbon
 (1)
(1)Biosensor
Escherichia coli HB101 was marked with the lux CDABE genes, originally isolated from Vibrio fischeri, using the multicopy plasmid pUCD607. The construction origin of this sensor is fully explained in Reid et al. [28]. It was stored as freeze-dried cultures (—20°C), which had been prepared according to standard laboratory protocols [16], and resuscitated for 1 hr in 10-ml 0.1M KCl before use. Cell suspensions (100 μL) of the biosensor were pipetted into 2-ml cuvettes containing the soil pore water (900 μL) and mixed for 5 s, at 15-s intervals between each sample. The final cell concentrations were between 2 × 107 colony forming units/ml and 1 × 107 colony forming units/ml. Luminescence was measured using a 1-s assay on a Bio-Orbit 1253 luminometer (Labtech International, Uckfield, UK). All assays were carried out in triplicate. Values were expressed as percentage of the luminescence in the control soil after 80-min exposure to the test solution.
Statistical analyses
The concentrations of metals in soil solution ranged by two to more than three orders of magnitude, and were transformed logarithmically before statistical analysis. Sigmaplot 8.0 (SPSS, Chicago, IL, USA) was used to fit either linear or Gompertz functions to the dose-response curves of bioassay results and the metal concentrations in soil solution. Genstat 6.0 (VSN International, Hemel Hempstead, UK) was used to test the significance of the regression models, using analysis of variance. Regression coefficients quoted are adjusted r2 values (r2 adj) and represent the percentage of the variance accounted for by the regression model.
RESULTS AND DISCUSSION
All the metal-amended microcosm soils had the same quantity of metals added to them. Total soil metal concentrations in the control soils (Table 1) were typical of uncontaminated soils [17]. No relationship was found between the ambient (control) total metal concentrations in control soils or total concentrations in the amended soils and the bioassay effects. However, the soluble metal concentrations in soil pore water varied greatly between soils. The differing chemical and physical properties of different soils affect the binding of metal ions on the soil components and alter the bioavailability.
The selection of the range of metal concentrations added in this study was close to current regulatory levels [18], which for some metals may be below concentrations that will cause any toxic effect. For example, soluble lead concentrations generally were low and, in most cases, at or below the detection limit due to the strong sorption of Pb in soil [29, 30].
At days 11 and 818 after amendment, the concentrations of Zn, Cd, Cu, and Ni in the soil solution correlated strongly with each other (r2 = 0.86–0.99). Any responses recorded in the bioassays theoretically could be due to a combination of these metals rather than any one of them. However, previous knowledge exists concerning the threshold metal concentrations that affect many of these assays, as discussed below.
It has been reported that both soil microbial biomass and MPN of Rhizobium are unaffected at the Cd and Ni levels used in this study [2, 8]. As a consequence of these observations, the data for these assays have been presented using the most likely causal agents (Zn and Cu). Little information is in the literature on the relative toxicity of metals in soils to Biolog responses, so the same presentation was followed as for Rhizobium and soil microbial biomass. The known effective concentration causing 50% reduction in luminescence (EC50) values of biosensors in aqueous systems suggest that the doses of Cd, Ni, and Cu encountered in this study were unlikely to cause a significant response [12, 21]. Accordingly, results of the biosensor are shown here only in relation to Zn concentrations in solution.
For the Woburn soil, the initial values for MPN rhizobia were similar to those reported by Chaudri et al. [2] demonstrating the repeatability of this technique. The MPN of rhizobia in control soils at day 11 were between 1.5 and 4.5 log10 units (data not shown), and remained similar in control microcosms after 818 d (Fig. 1), showing that storage alone did not reduce the number of rhizobia present. After only 11d, no effect of metals on MPN results could be seen (data not shown). However, after 818 d, rhizobial numbers decreased by approximately three orders of magnitude with increasing soluble concentrations of Zn and Cu (r2 adj for Zn = 0.38 and Cu = 0.54; both p < 0.001; Fig. 1). It has been shown previously in laboratory experiments that a long time is needed for metal toxicity to affect Rhizobium. Chaudri et al. [2] found no response after 2 months; however, after 18 months a decrease in rhizobial numbers was observed and, in the case of the most-contaminated soils, complete extinction of rhizobia.
Chaudri et al. [5] also found, in a long-term field experiment in which the soil had been contaminated with Zn or Cu from sewage sludge, that for R. leguminosarum biovar trifolii the lowest-observed-effect concentration of Zn in soil pore water was 0.6 mg/L and the EC50 was 7 mg/L. Their lowest-observed-effect concentration for Cu was 0.4 mg/L, but the EC50 could not be determined in their experiments because the maximum soluble Cu concentration was only approximately 0.7 mg/L. Although the effect of a single metal cannot be distinguished from the others in our experiment, in the context of the data in Chaudri et al. [5], it is unlikely that Cu was the causal agent of the reduction in MPN values because the highest soluble Cu concentration here was only 0.45 mg/L.
Figure 2 shows the relationship between biomass C and soil pore water metal concentrations on days 11 and 818. It can be seen that a dose-response relationship becomes more evident with time (day 11 r2 adj for Zn = 0.08 and Cu = 0.01, both not significant; day 818 r2 adj Zn = 0.41 p < 0.001 and Cu = 0.29, p < 0.01). Per unit soluble Zn or Cu, a greater reduction in biomass occurred at day 818 than at day 11, indicating that the mode of toxicity induced by the metals is cumulative, not immediate. It has been observed, however, that in metal-contaminated soils more energy is used for cell maintenance and hence less for growth, resulting in a higher metabolic quotient [31-34]. Therefore the population size gradually decreases in the presence of metal contamination, as cells die and less new cells are produced to replace them. This observation also means that a measurement such as microbial biomass likely is to be of more use than respiration measurements alone.
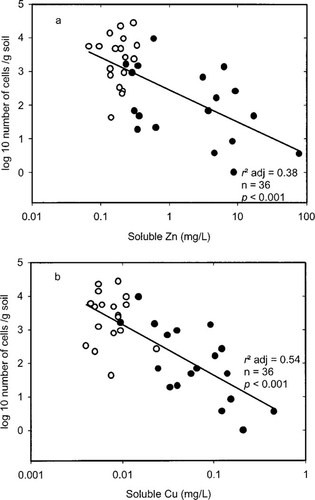
The effect of soluble concentrations of (a) Zn and (b) Cu on the population of Rhizobium leguminosarum biovar trifolii in the microcosm soils 818 d after amendment with metal nitrates. Control microcosms (○), metal-amended microcosms (•).
Figure 3 shows the number of substrates used in Biolog Ecoplates against soluble metal concentrations on days 11 and 818. The number of substrates used was insensitive to soluble metal concentration (day 11 r2 adj for Zn = 0.14 p < 0.01 and Cu = 0.09, p < 0.05 d 818 r2 adj Zn = 0.13 p < 0.05 and Cu = 0.07, not significant). This indicates that the functional diversity of the microbial community does not change when in contact with this concentration of added metals. Equally, it may be that Ecoplates do not show the change effectively, as only 31 carbon substrates are used and they have not been selected specifically to mimic the soil environment. Konopka et al. [9] concluded that the reliance on culturability was the main drawback of using Biolog to produce substrate utilization profiles. Other authors have concluded that Biolog profiling is a valuable comparative tool, as long as its limitations are understood and considered carefully [35, 36].
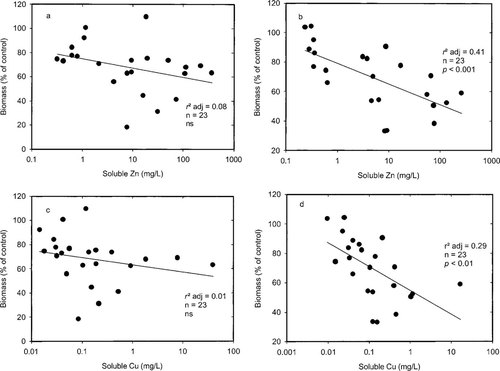
The effect of soluble Zn concentrations after 11 or 818 d (a,b), or soluble Cu concentrations after 11 or 818 d (c,d) on microbial biomass C in the microcosm soils following amendment with metal nitrates.
The bioluminescence-based biosensor responds to the bio-available metal in the soil solution [15]. Unlike the other bio-assays, the biosensor responded similarly to soil solution metal concentrations at 67 and 818 d, indicating that no change in metal speciation in solution had occurred. Data for these two measurement times were combined (Fig. 4) and one dose-response curve fitted all soils (r2 adj = 0.81, p < 0.001), and gave an EC50 value for soil solution Zn of 13 mg/L for E. coli.
For MPN rhizobia and biomass C to be useful in the assessment of metal-contaminated land, an uncontaminated control soil, which is the same soil type as the contaminated soil, must be present in the surrounding area. If a control is available, both MPN rhizobia and biomass C would be good indicators of bioavailable metals and their toxicity to the microbial community, whether the land is contaminated with either one or many metals. The reduction in both of these bio-assays compared to the control indicates large impacts on the whole microbial community (biomass C) or an important species of bacteria (R. leguminosarum bv. trifolii). On the other hand, the biosensor responds to the acute toxicity of the solution phase of the soil. No marked decrease with time was found in the Zn or Cu in soil solution (Figs. Fig. 2., Fig. 3.). Despite this, the microbial biomass and rhizobial MPNs were impacted significantly at the end of the incubation but not near the start, indicating cumulative effects. The Biolog results were found to be independent either of the chemical data or the other biological data. The number of substrates utilized decreased with time (Fig. 3) and this may indicate that the impact of the incubation was greater than that of the treatments, as reported by Bundy et al. [37].
CONCLUSION
For the soils tested, a range of microbial assays was found to be more informative than a single test. To fully understand the value of these different microbial techniques in appraising metal contaminants in soils, we need to evaluate them in different ways. Not all techniques were applicable to the full suite of soil conditions: Most important was that the Rhizobium MPN and Biolog methods could not be applied to strongly acidic soils. The other tests should be applicable to a wider range of soils. To fully justify the key tests for ecotoxicity testing in soils, we must select the assays on the basis of environmental relevance, applicability to a wide range of soils and pollutants, and ease of interpretation of response. The bioassays used here: Rhizobium survival in soils, the size of the soil microbial biomass, the Biolog pattern of responses, and a single-species biosensor represent different receptor communities that have different diversity and tolerance. Added to that are differences in the mode of exposure (in situ, ex situ). Biolog is an ex situ growth of bacteria extracted from soil, under nonexposed conditions. Biosensors reflect the planktonic bacteria in soil pore water and, therefore, are exposed only to the solution-phase pollutants; however, the Rhi-zobium MPN and the microbial biomass measurements reflect whole soil exposure pathways, and these showed increasing toxicity over time. This has important implications for soil monitoring and for soil toxicological assessment with respect to metals. It also may prevent any direct comparisons between the different types of test as assessment tools.
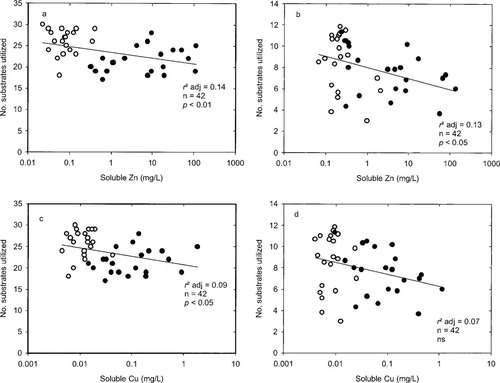
The effect of soluble Zn concentrations after 11 or 818 d (a,b), or soluble Cu concentrations after 11 or 818 d (c,d) on the number of substrates used in Biolog Ecoplates (Hayward, CA, USA) following amendment of the microcosm soils with metal nitrates. Control microcosms (○), metal-amended microcosms (•).
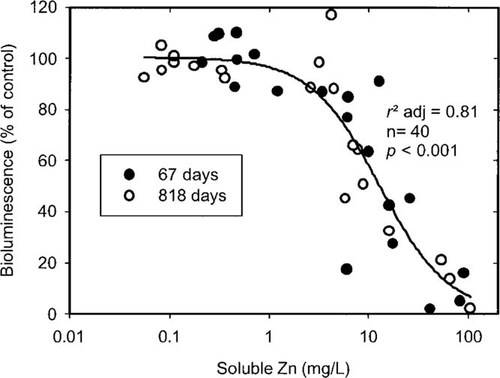
The effect of soluble Zn concentration on the luminescence response of the Escherichia coli biosensor 67 d and 818 d after amendment of the microcosm soils with metal nitrates.
Acknowledgements
This work was funded by Grant T06499 from the United Kingdom Biotechnology and Biological Sciences Research Council.




