Phylogeography of the Bent-Toed Geckos (Squamata: Gekkonidae) Reveals Complex Diversification Patterns Linked to the Orogenic History of the Himalayas
Abstract
The evolutionary history of the Himalayan biota has been shaped by geological and climatic changes over time. The Himalayan bent-toed geckos, Cyrtodactylus (Squamata: Gekkonidae), is an ideal group to study the phylogeography of the Himalayas due to its early origin, wide distribution, and ongoing diversification across its range. However, the specific impact of geological events on the diversification of Central Himalayan Cyrtodactylus has not been fully investigated. In this study, we sampled Cyrtodactylus from the Central Himalayan region and analyzed concatenated mitochondrial (NADH dehydrogenase 2, 1038 bp) and nuclear (phosducin [PDC], 393 bp, and recombination activating gene, 861 bp) DNA sequences totaling 2292 bp. We complemented these by the sequences from the Himalayan and the Indo-Burma regions. Phylogenetic analyses revealed that the Cyrtodactylus samples from the Himalayan region are grouped into four major groups: C. lawderanus group, C. fasciolatus group, C. peguensis group, and C. khasiensis group. The geckos sampled in this study from the Central Himalayas belonged to the C. fasciolatus and C. khasiensis groups. Our results indicate that Cyrtodactylus underwent radiation events from the early Eocene to the late Pleistocene, with a relatively constant rate of divergence but a significant deviation from a constant rate of lineage accumulation. These findings establish a complex relationship between gecko diversification and the orogenic process of the Himalayas, which created a diverse landscape, global climate changes that led to different environments, intermittent arid conditions, shifting south Asian monsoon patterns, and the evolution of rivers. These factors facilitated allopatric speciation in the Himalayan region. Our results support a west to east diversification pattern within the range of Cyrtodactylus. Further extensive sampling and integrated genetic and ecological analyses are warranted to understand the evolution of the least studied herpetofauna of the Central Himalayan region.
1. Introduction
Geological and climatic changes during the earth’s history have shaped biodiversity and left distinct imprints of historical biogeography. The collision of the Eurasia and Indian plates, a prominent geological event that occurred ~50 million years ago, had a major impact on Asian biodiversity by linking the previously isolated biota of the Indian Plate with that of Eurasia [1, 2]. This geological shift allowed extensive exchange of fauna between Peninsular India and Eurasia since the Eocene. Molecular evidence supports biotic links between Peninsular India and mainland Asia during the Eocene–Oligocene, with dispersal both into and out of the Indian subcontinent, including the Himalayan region [3, 4]. Additionally, the uplift of the Himalayas and the intensification of the Asian monsoon system have been key drivers of species diversification in the region surrounding the Tibetan Plateau and the Himalayas, facilitating uplift-driven in-situ diversification and impacting biotic interchanges [5, 6]. Molecular analyses of taxa represented by a large number of species and distributed across a wide spatial extent can help to elucidate past geological and climatic events and their impact on life forms.
The genus Cyrtodactylus (Squamata: Gekkonidae) is distributed across South and Southeast Asia and extends into Melanesia, occupying diverse and endangered environments, including the trans-Himalayan Tibetan Plateau and the Himalayan regions of India, Nepal, and Pakistan [7–12]. Their evolutionary history is complicated by their exceptional dispersal ability, cryptic nature, and spread across diverse terrains, providing insights into the geological and ecological factors influencing their phylogenetic structure and diversity [10, 11, 13]. The unique evolutionary history of the Himalayan biota has been shaped by geological and climatic changes over time. In addition, the isolation of the region by geographical features, such as mountains and rivers is likely to have acted as an important driver of biological diversification in these areas [7, 14–17]. Thus, Cyrtodactylus is an ideal group for understanding the phylo-biogeography of the Himalayan region due to its early origin and continuous diversification and diversion across its range over time.
Their evolutionary distribution and the continuous landscape changes have triggered the evolutionary divergence of Cyrtodactylus in the “proto-Himalayan” region [12]. Major divergence events likely occurred between 62 and 47.7 million years ago (mya) during the early Eocene [7, 9, 13] and resulted in west to east radiation of species from South Asia to Melanesia [12]. These early divergences are thought to be a consequence of landscape changes during the continuous compression of Neotethyan island arcs, such as the Kohistan Arc [7, 10–12, 18, 19]. Subsequent divergence due to landscape and climate changes, in particular the uplift of the Himalayas, the impact of monsoons causing soil erosion, and prolonged aridification, has led to the clustering of species and the formation of distinct groups [7, 12]. However, the precise impact of these geological events on the diversification of Central Himalayan Cyrtodactylus from the early Eocene to the present remains uncertain. This is due to the limited number of Central Himalayan species included in the sampling and the fact that recently discovered species and analyses of diversification rates and ancestral ranges have not yet been fully incorporated [7, 8].
This study aimed to understand the evolutionary dynamics of the bent-toed geckos in the Himalayas in the context of past geological and climatic changes. By analyzing additional samples of the genus Cyrtodactylus, the study provides valuable insights into the evolutionary past of the genus and offers a comprehensive perspective on the evolution of species in the Himalayan region.
2. Materials and Methods
2.1. Sampling, DNA Extraction, PCR Amplification, and Sequencing
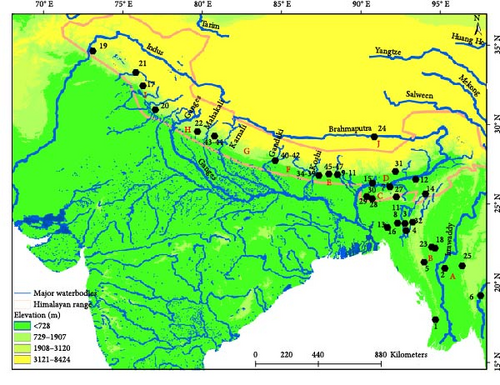
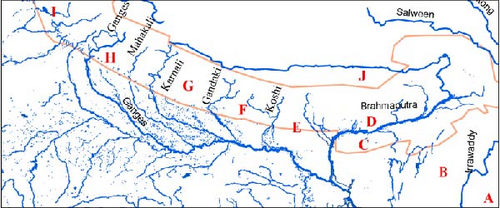
To address the gap in the phylogeographical analysis of bent-toed geckos (Cyrotodactylus) from the Central Himalayas [7, 8], we conducted field surveys and sampling in Koshi River, Gandaki River, and Karnali–Mahakali River basins of Nepal (Figure 1). During these surveys, we collected 18 gecko specimens from the study area. We sub-sampled 14 specimens for molecular analysis to ensure representation from all sampling localities. This effort aims to improve our understanding of the evolutionary dynamics of Cyrtodactylus in the region, building on previous studies that have highlighted the need for broader sampling and analysis to reveal the complex biogeographical patterns in the Himalayas.
| Site | Species | Museum number | Area | Locality | ND2 | RAG1 | PDC |
|---|---|---|---|---|---|---|---|
| 1. | Cyrtodactylus ayeyarwadyensis | CAS 216459 | West of Irrawaddy River (B) | Myanmar, Rakhine State, Than Dawe District | JX440526 | JX440685 | JX440634 |
| 2. | Cyrtodactylus brevidactylus | CAS 214104 | East of Irrawaddy River (A) | Myanmar, Mandalay, Popa Mountain | JX440527 | JX440687 | JX440636 |
| 3. | Cyrtodactylus aaronbaueri | MZMU2015 | West of Irrawaddy River (B) | India, Mizoram, Durtlang North | MW596520 | — | — |
| 4. | Cyrtodactylus bengkhuaiai | MZMU1985 | West of Irrawaddy River (B) | India, Mizoram, Sailam Community Reserved | MW596516 | — | — |
| 5. | Cyrtodactylus gansi | CAS 22412 | West of Irrawaddy River (B) | Myanmar, Chin State (Min) Dat Township | JX440537 | JX440697 | JX440646 |
| 6. | Cyrtodactylus myintkyawthura | USNM 559805 | East of Irrawaddy River (A) | Myanmar, Mandalay Division, Popa Mountain Peak | JX440536 | JX440696 | JX440645 |
| 7. | Cyrtodactylus guwahatiensis | BNHS 2146 | West of Irrawaddy River (B) | India, Assam, Guwahati District | KM255194 | KM255135 | KM255160 |
| 8. | Cyrtodactylus montanus | CES10/1211 | West of Irrawaddy River (B) | India, Tripura, North District, Phuldungsei | KM255200 | KM255138 | KM255163 |
| 9. | Cyrtodactylus gubernatoris | CES09/1197 | West of Brahmaputra and east of Koshi (E) | India, Sikkim, Singtam | KM255181 | KM255139 | KM255165 |
| 10. | Cyrtodactylus bhupathyi | CES10/1235 | West of Brahmaputra and east of Koshi (E) | India, west Bengal, Kalimpong | KM255204 | KM255123 | KM255147 |
| 11. | Cyrtodactylus jaintiaensis | BNHS 2248 | South of Brahmaputra (C) | India, Assam, Meghalaya, Jaintia Hills District | KM255195 | — | — |
| 12. | Cyrtodactylus kazirangaensis | BNHS 2147 | South of Brahmaputra (C) | India, Assam, Golaghat District, Kohora | KM255170 | KM255136 | KM255161 |
| 13. | Cyrtodactylus khasiensis | BNHS 2249 | West of Irrawaddy River (B) | India, Meghalaya, East Khasi District | KM255188 | KM255127 | KM255152 |
| 14. | Cyrtodactylus nagalandensis | BNHS 2253 | South of Brahmaputra (C) | India, Nagaland, Kohima District | KM255199 | KM255137 | KM255162 |
| 15. | Cyrtodactylus septentrionalis | BNHS 1989 | North of Brahmaputra (D) | India, Assam, Bongaigaon District | MH971164 | KM255133 | KM255158 |
| 16. | Cyrtodactylus tripuraensis | BNHS 2238 | West of Irrawaddy River (B) | India, Tripura, West District | KM255183 | KM255142 | KM255167 |
| 17. | Cyrtodactylus chamba | CES11/1291 | East of Indus and west of Ganges (I) | India, Himanchal Pradesh, Chamba District | KM255191 | KM255134 | KM255159 |
| 18. | Cyrtodactylus annandalei | CAS 215722 | West of Irrawaddy River (B) | Myanmar, Sagaing Division, Alaungdawa Kathapa National Park | JX440524 | JX440683 | JX440633 |
| 19. | Cyrtodactylus battalensis | PMNH 2301 | East of Indus and west of Ganges (I) | Pakistan, NWFP, Battagram City | KC151983 | KC152035 | KC152007 |
| 20. | Cyrtodactylus fasciolatus | CES11/1337 | East of Indus and west of Ganges (I) | India, Himachal Pradesh, Shimla District, Nr. Subathu | KM255184 | KM255120 | KM255143 |
| 21. | Cyrtodactylus himalayanus | CES11/1297 | East of Indus and west of Ganges (I) | India, Jammu and Kashmir, Doda District, Bhaderwah | KM255173 | KM255124 | KM255149 |
| 22. | Cyrtodactylus lawderanus | CES11/1343 | East of Ganges and west of Mahakali (H) | India, Uttarakhand, Almora District, Almora | KM255189 | KM255128 | KM255154 |
| 23. | Cyrtodactylus slowinskii | CAS 210205 | West of Irrawaddy River (B) | Myanmar, Sagaing Division, Htamanthi Wildlife Sanctuary | JX440559 | JX440719 | — |
| 24. | Cyrtodactylus tibetanus | MVZ 233251 | Trans-Himalaya (J) | Tibet, Lhasa, 3 km WNW of Potala Palace | JX440561 | JX440722 | — |
| 25. | Cyrtodactylus chrysopylos | CAS226141 | East of Irrawaddy River (A) | Myanmar, Shan State, Ywa Ngan Township | JX440531 | JX440690 | JX440639 |
| 27. | Cyrtodactylus urbanus | VR/ERS/ZSI/688 | South of Brahmaputra (C) | India, Assam, Kamrup (M) District, Guwahati, Basishta | MN911174 | — | — |
| 28. | Cyrtodactylus karsticola | MZMU2153 | South of Brahmaputra (C) | India, Meghalaya, South Garo Hills, Siju | MW596513 | — | — |
| 29. | Cyrtodactylus agarwali | MZMU2158 | South of Brahmaputra (C) | India, Meghalaya, South Garo Hills, Siju | MW596515 | — | — |
| 30. | Cyrtodactylus bapme | BNHS 2754 | South of Brahmaputra (C) | India, Meghalaya State, West Garo Hills Dist., Jangrapara Village | MW367438.1 | — | — |
| 31. | Cyrtodactylus kamengensis | BNHS 3113 | North of Brahmaputra (D) | Shergaon, West Kameng District, Arunachal Pradesh | OM023868 | — | — |
| 32. | Cyrtodactylus ngopensis | MZMU2553 | West of Irrawaddy River (B) | Mizoram, India | OM912605 | — | — |
| 33. | Hemidactylus frenatus | — | — | India | HM559629 | EU108534 | EU268328 |
| 34. | Cyrtodactylus CDZMTU501 sp.nov.1 | CDZMTU501 | West of Brahmaputra and east of Koshi (E) | Dharan Base Camp, Eastern Nepal | This study | This study | This study |
| 35. | Cyrtodactylus CDZMTU501 sp.nov.1 | CDZMTU502 | West of Brahmaputra and east of Koshi (E) | Dharan Base Camp, Eastern Nepal | This study | This study | This study |
| 36. | Cyrtodactylus CDZMTU501 sp.nov.1 | CDZMTU503 | West of Brahmaputra and east of Koshi (E) | Dharan Base Camp, Eastern Nepal | This study | This study | This study |
| 37. | Cyrtodactylus CDZMTU501 sp.nov.1 | CDZMTU504 | West of Brahmaputra and east of Koshi (E) | Dharan Base Camp, Eastern Nepal | This study | This study | This study |
| 38. | Cyrtodactylus CDZMTU501 sp.nov.1 | CDZMTU505 | West of Brahmaputra and east of Koshi (E) | Dharan Base Camp, Eastern Nepal | This study | This study | This study |
| 39. | Cyrtodactylus CDZMTU501 sp.nov.1 | CDZMTU506 | West of Brahmaputra and east of Koshi (E) | Dharan Base Camp, Eastern Nepal | This study | This study | This study |
| 40. | Cyrtodactylus CDZMTU507 sp.nov.2 | CDZMTU507 | West of Koshi and east of Gandaki (F) | Chitwan, Central Nepal | This study | This study | This study |
| 41. | Cyrtodactylus CDZMTU508 sp.nov.2 | CDZMTU508 | West of Koshi and east of Gandaki (F) | Chitwan, Central Nepal | This study | This study | This study |
| 42. | Cyrtodactylus CDZMTU509 sp.nov.2 | CDZMTU509 | West of Koshi and east of Gandaki (F) | Chitwan, Central Nepal | This study | This study | This study |
| 43. | Cyrtodactylus nepalensis | CDZMTU510 | West of Gandaki and east of Mahakali (G) | Dadeldhura, Western Nepal | This study | This study | This study |
| 44. | Cyrtodactylus nepalensis | CDZMTU511 | West of Karnali and east of Mahakali (G) | Dadeldhura, Western Nepal | This study | This study | This study |
| 45. | Cyrtodactylus martinstolii | CDZMTU512 | West of Brahmaputra and east of Koshi (E) | Illam, Eastern Nepal | This study | This study | This study |
| 46. | Cyrtodactylus martinstolii | CDZMTU513 | West of Brahmaputra and east of Koshi (E) | Illam, Eastern Nepal | This study | This study | This study |
| 47. | Cyrtodactylus martinstolii | CDZMTU514 | West of Brahmaputra and east of Koshi (E) | Illam, Eastern Nepal | This study | This study | This study |
- Note: The serial numbers refer to the locality numbers in Figure 1. The block letters in parentheses under the heading “Area” indicate the 10 broad geographical areas defined for ancestral range reconstruction. BNHS, Bombay Natural History Society, Mumbai; CAS, California Academy of Sciences, San Francisco; CDZMTU, Central Department of Zoology Museum, Tribhuvan University, Kathmandu; CES, Centre for Ecological Sciences, Bangalore; MVZ, Museum of Vertebrate Zoology, Berkeley; MZMU, Departmental Museum of Zoology, Mizoram University, Aizawl; PMNH, Pakistan Museum of Natural History, Islamabad; USNM, United States National Museum, Washington; VR/ERS/ZSI, National Centre for Biological Sciences, Bangalore (NCBS) and National Zoological Collection, maintained by the North Eastern Regional Centre, Zoological Survey of India, Shillong.
- aPreviously considered Cyrtodactylus feae by Agarwal et al. [7, 8] and Wood et al. [12], later revised to Cyrtodactylus myintkyawthuri by Grismer et al. [20, 21].
Genomic DNA was extracted from 2.5 mg of liver and/or skeletal muscle samples preserved in 100% ethanol using the QIAmp DNA Mini Kit (Qiagen, Germany) following the protocols of the manufacturer. A mitochondrial gene, NADH dehydrogenase 2 (ND2, 1038 bp), and partial sequences of the two nuclear genes, phosducin (PDC, 393 bp) and recombination activating gene (RAG1, 861 bp), were amplified using the primer pairs following Bauer et al. [16] (Table S1). The PCR reaction volume contained 25 μl with 12.5 μl Power Taq PCR Master Mix (BioTeke, Beijing, China), 0.5 μl each of forward and reverse primers, 10.5 μl deionized distilled water, and 1 μl template DNA. The PCR thermal cycles included an initial denaturation at 95°C for 5 min and 35 cycles of denaturation at 94°C for 30 s, annealing at a specific temperature for 30 s (Table S1), and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. DNA extraction and PCR were performed in the molecular laboratory at the Central Department of Zoology, Tribhuvan University.
Amplicons were tested using electrophoresis on 1.5% agarose gels. The successful amplicons were sequenced using a Big Dye Terminator Cycle Sequencing Kit (Invitrogen, USA) and then analyzed on an ABI Prism 3700 Automated Sequencer (Applied Biosystems, USA). Both strands of successful amplicons were sequenced using the same primer pairs as in PCR. The sequences from the two strands were assembled into a single FASTA file using the SeqMan tool in the DNASTAR Lasergene package [22].
The homologous sequences for the three genes under study, belonging to the genus Cyrtodactylus from the Himalayan and Indo-Malayan regions, were retrieved from GenBank (Table 1). Additionally, corresponding sequences from Hemidactylus frenatus were used as an outgroup in phylogenetic analysis. Sequences were aligned using the ClustalW algorithm in MEGA 11 [23]. The alignments for the three genes of each species were then concatenated using PhyloSuite v1.2.3 [24].
2.2. Phylogenetic Analysis
Phylogenetic relationships among geckos were analyzed using 46 homologous sequences, including 33 from the Himalayan region and 13 from the Indo-Burma region. The Indo-Burma specimens were included to understand the mixed diversification patterns of Cyrtodactylus species. The molecular phylogeny of Himalayan Cyrtodactylus was reconstructed using two alignments i) mitochondrial ND2 (1038 bp), and ii) concatenated mitochondrial and nuclear data (ND2 + PDC + RAG1, 2292 bp), with maximum likelihood (ML) and Bayesian inference (BI) methods. The best-fit partitioning scheme for the alignment was determined using PartitionFinder2 v2.1.1 [25], with a greedy algorithm and BIC criterion. ModelFinder v2.2.0 [26] was used to select the best-fit substitution model (edge-linked) using the BIC criterion (GTR + G + I for ND2 alignment; see Table S2 for the concatenated alignment). The Phylip format file for the alignment generated using Geneious Prime 2024.0.5 [27] was used for ML analysis. Partitioned ML analyses were conducted using RAxML 8.2.12 implemented in RAxML GUI v2.0.10 [28, 29] with 1000 rapid bootstraps.
For the BI analyses, the input “xml” file was prepared in BEAUti v10.5.0. The input file was run in BEAST2 v10.5.0 [30] with two independent Markov Chain Monte Carlo (MCMC) chains, each with 20,000,000 generations. Samples were taken every 1000 generations, with the first 10% of total generations discarded as burn-in. A <0.01 deviation in the split frequencies between the two independent runs ensured convergence of the two MCMC chains. Convergence was further assessed using TRACER v.1.6 [31], with an effective sample size (ESS) >200. The single maximum clade credibility (MCC) tree was generated using TreeAnnotator v10.5.0. Both the ML and BI phylogenetic trees were visualized using FigTree v1.4.4 [32]. The tree topologies of the ML and BI trees were compared using phylo.io (https://phylo.io/) [33].
Pairwise genetic distances among Cyrtodactylus species were calculated using the Kimura-2-parameter based on ND2 sequences (1038 bp) without the pairwise deletion option using MEGA11 [23].
2.3. Divergence Dating and Diversification Analysis
Divergence dates among the study taxa were estimated using concatenated mitochondrial (ND2) and nuclear (RAG1 and PDC) alignments with the partitioning schemes and substitution models determined above. The input file was generated in BEAUti v.1.10.4 with a lognormal relaxed clock, unlinked site models, linked trees and clock models, and a Yule prior [34]. MCMC chains were run for 20,000,000 generations, with samples taken every 1000 generations. Two priors previously employed in studies were utilized for calibrated analyses: one for the divergence of H. frenatus from Cyrtodactylus ancestors around 64 million years ago (73–56 mya) [12], and another for the split of C. tibetanus from the rest of the Cyrtodactylus group at 55 mya (64–47 mya) [7, 10, 11, 35]. The log files generated by BEAST were examined and evaluated in Tracer v. 1.7.2 [32] to ensure that the ESSs were greater than 200 for all parameters [36]. The first 10% of generations were removed as burn-in, and the MCC tree was generated using TreeAnnotater v1.8.4. FigTree v1.4.4 [32] was used to visualize the tree.
The resulting MCC BI tree with 46 tips and 45 internal nodes, excluding the outgroup, was used for lineage-through-time (LTT) analysis. A LTT analysis was performed using the “ltt” function in the “phytools” and “ape” packages [37] in R version 4.1.1 [38]. The gamma statistic (γ) was calculated to test for deviations from a constant diversification rate. To assess the significance of the observed gamma statistics, we simulated 100 phylogenetic trees under a pure-birth model using the “pbtree” function. The simulated trees were then analyzed to generate a null distribution of gamma values. We calculated the Type I error rate by determining the proportion of simulated p-values less than or equal to 0.05. Multiple trees were calculated to resolve the uncertainty and provide a more robust conclusion using the Laser package [39]. Coalescent trees were used to model the genealogical history of samples from a population, focusing on how lineages coalesce over time. We tested whether coalescent trees using the “rcoal” function built in the ape package deviated from a constant rate of lineage accumulation from the simulated pure-birth trees for incomplete linkage by comparing the observed gamma statistic with the distribution of gamma values [40].
2.4. Ancestral Range Reconstruction
To estimate the historical biogeography of Cyrtodactylus in the Himalayan region through ancestral area reconstruction, the best evolutionary model (BAYAREALIKE + J) was determined by the BioGeoBEARS v1.1.1 [41, 42] R package based on the highest AICc_wt value. We ran the analysis using BioGeoBEARS v1.1.1 [41, 42], which is used in Reconstruct Ancestral State in Phylogenies (RASP v4.4) as described by Yu et al. [43, 44]. The analysis used the MCMC BEAST tree chains for 20,000,000 generations, with logging occurring every 1000 generations. The first 2000 trees (10% burn-in) were discarded, and the remaining 19801 trees were used for binary tree analysis. The analysis used the MCC BI tree generated using TreeAnnotator v10.5.0 as the consensus tree in user-specified trees. We set 100 random trees to be used in the analysis and a maximum geographic area of 2 [44]. The species in the study were grouped into 10 broad areas based on geological features such as distance to each other, rivers, and mountains, with a focus on the biogeography of the Central Himalayas. They were defined as (A) east of Irrawaddy River, (B) west of Irrawaddy River, (C) south of Brahmaputra, (D) north of Brahmaputra, (E) west of Brahmaputra and east of Koshi, (F) west of Koshi and east of Gandaki, (G) west of Gandaki and east of Mahakali, (H) east of Ganges and west of Mahakali, (I) east of Indus and west of Ganges, and (J) Trans-Himalaya. Areas A–E and area J were considered based on the findings of Agrawal et al. [7, 8]. For the Central Himalaya and Western Himalaya, major rivers were considered as possible dispersal barriers due to their early onset of evolution approximately 30 mya during the orogenesis of the Himalayas [45]. The main objectives of the analysis were to determine (1) dispersal, vicariance, and extinction events in the Himalayan region; (2) regions with higher dispersal rates with frequent exchange of species, resulting in greater gene flow and more similar biotic communities; (3) regions with lower dispersal rates that may experience greater isolation, potentially leading to increased differentiation or speciation over time; and (4) potential pathways for species dispersal within or across the Himalayan region. Further, to understand the role of environmental variables in the distribution of bent-toed geckos, the difference in the number of bent-toed gecko species occurrences with elevation, precipitation, and annual average temperature was tested for statistical significance using analysis of variance (ANOVA).
3. Results
3.1. Phylogenetic Relationships Among the Bent-Toed Geckos
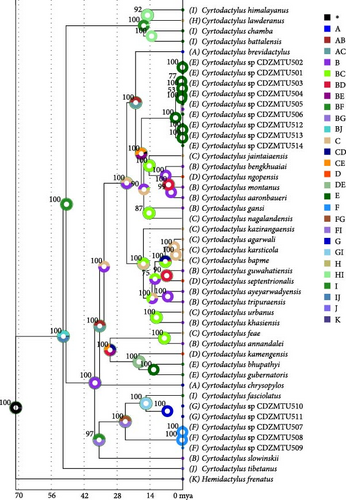
The ML and BI trees based on the mitochondrial ND2 gene (1038 bp) yielded similar tree topology with strong bootstrap/BPP support (Figure 2 and Figure S1), largely consistent with that of Grismer et al. [10, 11] regarding previously analyzed species. The trees indicated that Cyrtodactylus samples from the Himalayan region form four major clades: the lawderanus group, the fasciolatus group, the peguensis group, and the khasiensis group as originally defined by Grismer et al. [10, 11]. In addition, phylogenetic trees based on the concatenated mtDNA and nuclear DNA (Figure S2) showed similar tree topologies to those based on mtDNA alone.

Sequences from the Western Himalaya (west of the Ganges River) of C. lawderanus, C. battalensis, C. chamba, and C. himalayanus formed a monophyletic group, referred to as the C. lawderanus group. Phylogenetic analysis revealed that C. fasciolatus, originally described from the Western Himalaya (east of the Ganges River), forms a distinct clade with sequences from the Central Himalayan region (CDZMTU507–CDZMTU5011), referred to as the C. fasciolatus group. Similarly, sequences from eastern Nepal (CDZMTU501–CDZMTU506 and CDZMTU512–CDZMTU514) exhibited monophyletic origin.
The C. khasiensis group is divided into four major subgroups: (1) lower-elevation species averaging 240 m, consisting of Northeast Indian/Himalayan species and species south of the Brahmaputra River Basin and the Indo-Burma region; (2) higher-elevation species averaging 1254 m, including Indo-Burma and Northeast Indian/Himalayan species; (3) Northeastern Himalayan and Myanmar species; (4) Eastern Himalayan species from Nepal. In addition, the eastern Himalayan species C. kamagensis, C. bupathyi, and C. guberatoris are strongly monophyletic with C. peguensis.
3.2. Divergence Times of the Bent-Toed Geckos
The results of our BEAST analysis had an ESS of more than 200 for all parameters, indicating a robust and reliable estimate across the board. The study shows that Cyrtodactylus underwent radiation events from approximately 65 mya, spanning the early Eocene to the late Pleistocene fluctuations (Figure 3).
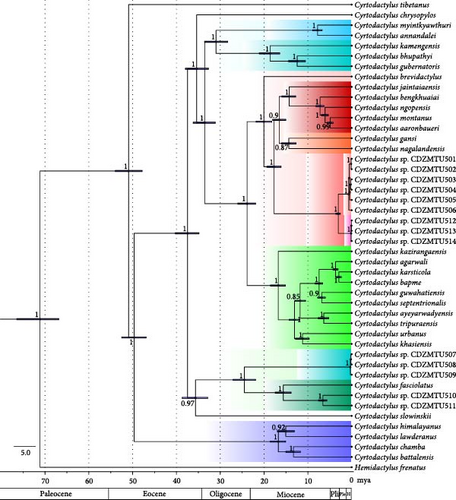
The results strongly suggest ancient radiation in the Old World, especially in the early proto-Himalayan region, forming a group with Cyrtodactylus and Palaearctic naked-toed geckos as Wood et al. [12], Agarwal et al. [7, 8], and Grismer et al. [10, 11]. C. tibetanus was the first gecko to diverge from the rest of the group, approximately 55 mya (95% HPD: 64–47). The Western Himalayan group (C. lawderanus) diverged from the Central Himalayan and Eastern Himalayan groups approximately 49 mya (95% HPD: 52.47–46.92) during the early Eocene. The ancestor of the Himalayan and Indo-Malayan species diverged around 37 mya (95% HPD: 40.24–34.85). The ancestor of C. peguensis from the Indo-Burma region and the ancestor of C. fasciolatus from the western Himalaya diverged around 34 mya (95% HPD: 37.95–32.71) and (95% HPD: 38.69–32.87), respectively. The ancestor of the C. peguensis group diverged from the ancestor of C. khasiensis around 33 mya (95% HPD: 36.17–31.12 mya). The peguensis group diverged from species in the Eastern Himalayas around 31 mya (95% HPD: 33.59–28.29 mya). Two simultaneous radiations occurred in the Central Himalayan region around 24 mya. The species of the fasciolatus group from Central Nepal diverged from the western congeners ~24 mya (95% HPD: 27.05–21.94), while the species of the khasiensis group from eastern Nepal, adapted to lower altitudes, diverged from those comparatively higher altitudes around the same time (95% HPD: 25.97–21.91). C. brevidactylus, found east of the Irrawaddy River Basin, split from the ancestor of the northern, comparatively highland Indo-Burman and Himalayan species around 20 mya (95% HPD: 21.81–18.25). The far eastern Himalayan species C. kamengensis separated from C. bhupathyi and C. gubernatoris around 19 mya (95% HPD: 21.06–16.4). Similarly, the highlander Indo-Burman species diverged from the eastern Himalayan species about 18 mya (95% HPD: 19.35–16.12). The relatively highland and lowland western Himalayan species diverged around 17 mya (95% HPD: 18.56–15.08). Cyrtodactylus nagalandensis from the eastern Himalayas and south of the Brahmaputra River Basin diverged from other highland Cyrtodactylus species of the Indo-Burma and Eastern Himalayan lineages about 17 mya (95% HPD: 18.06–15.04). The western Himalayan C. fasciolatus species and those from the east of the Mahakali River Basin diverged around 16 mya (95% HPD: 17.4–13.84). Around 15 mya (95% HPD: 15.92–12.77, 15.99–12.7, 16.97–13.07), C. jaintiaensis of the eastern Himalayas diverged from other Indo-Burma species. During this period (95% HPD: 15.99–12.7), C. nagalandensis from the eastern Himalayas also diverged from the species in Myanmar, and eastern and western C. lawderanus and C. himalayanus species from the Ganges River Basin diverged (95% HPD: 16.97–13.07 mya) from each other. Cyrtodactylus guwahatiensis and C. septentrionalis, species found north and south of the Brahmaputra River Basin, diverged from other species in the Nokrek National Park area around 7 mya (95% HPD: 8.32–6.61 mya). The two species diverged further around 6.5 mya (95% HPD: 7.79–6). Two molecular operational taxonomic units sampled in this study from eastern Nepal (CDZMTU501–CDZMTU506 and CDZMTU512–CDZMTU514) diverged around 3 mya (95% HPD: 3.68–2.58).
3.3. Diversification of the Bent-Toed Geckos in the Himalayas
The MCC phylogenetic tree, excluding the outgroup, containing 46 tips and 45 internal nodes, shows no significant deviation from a constant-rate speciation model, with a gamma value (γ) of 1.3416 and a p-value of 0.1797. This indicates a trend of recent speciation followed by a slowdown, but it was not statistically significant. However, analysis of simulated trees (n = 200) revealed that the proportion of trees with p-values ≤0.05 is only 1%, lower than the expected 5%. This suggests that the gamma test is conservative and less likely to falsely reject the null hypothesis of a constant rate of divergence. Nevertheless, visual inspection of the LTT plots suggested some major shifts in the number of lineages from the late 18 mya (Figure 4).
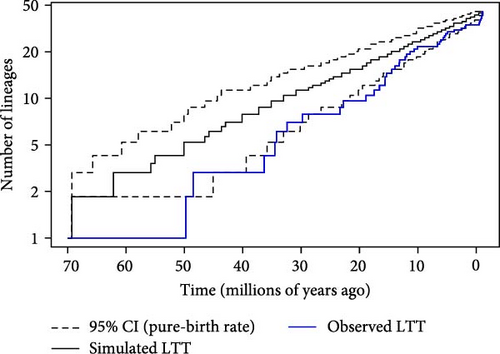
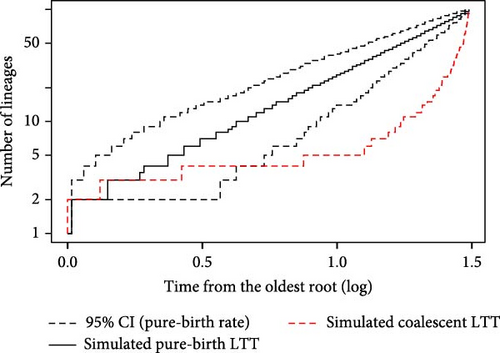
The gamma statistic (γ = 9.9591) with a p-value of <0.001 indicates a significant deviation from a constant rate of lineage accumulation. This high positive gamma value suggests a recent rapid increase in coalescent events, consistent with a recent population expansion. The LTT plot for the coalescent tree was superimposed on the 95% confidence intervals for the LTT curves of the pure-birth trees. The LTT plot of the coalescent tree (red, dashed) showed a significant deviation from the pure-birth expectation, further supporting the hypothesis of a recent lineage diversification.
3.4. Ancestral Range Reconstruction
A complex biogeographical history was recorded for all taxa studied, with a total of 47 evolutionary events. Of these events, 48.9% (n = 23) were dispersal events, 44.6% (n = 21) were vicariance events, and 6.3% (n = 3) were extinction events (Figure 5). Additionally, of the 45 nodes analyzed, 93.3% had a posterior probability of >0.95 (Figure 5), indicating strong support for these nodes. The major dispersal events occurred at 17.7–16.5, 7.4–7.2, 13.1–11.8, 24.5–20.0, 37.5–30.9, and 71.2–49.7 mya. Major vicariance events occurred between 15.6 and 14.2, 6.8–6.1, 16.8–11.2, and 71.2–50.9 mya.
The region west of the Irrawaddy River (B) stands out as a major source of species dispersal to other regions, with a total dispersal rate of 14.50. This area exhibits high movement values, such as the west of Irrawaddy River Basin (B) to South of Brahmaputra (C) with a dispersal rate of 3.5, suggesting frequent inter-area dispersal. The areas between the Koshi–Brahmaputra River basins and west of the Irrawaddy River basin have the highest number of speciation events with 9 and 8 events, respectively. This highlights those areas as major centers for Cyrtodactylus diversification and important speciation hotspots. The ancient western Himalayan lineages had substantial dispersal events to the west of Irrawaddy River Basin (B), as well as to the areas between the Gandaki–Mahakali (G) and the Ganges–Mahakali (H) with dispersal rates of 1, 1, and 0.5, respectively. However, taxa from the areas between the Ganges–Mahakali (H) and east of Irrawaddy River basin (A) exhibited no dispersal events to other areas in this analysis. These areas are likely to act as sinks, receiving species but not contributing to the dispersal of species, or they may be relatively isolated in terms of dispersal dynamics within the geography studied.
4. Discussion
Over the past 20 years, there has been a rapid increase in the description of Cyrtodactylus species, with the known diversity of the genus more than doubling since the beginning of the 21st century [12, 46]. This study included four molecular taxonomic units from the Central Himalayan region that had not previously been included in analyses, highlighting the previously incomplete understanding of the biogeography of Cyrtodactylus in the Himalayas. Existing knowledge of Cyrtodactylus lineages and phylogeny is limited, with many regions still lacking comprehensive phylogenetic resolution. This research, through phylogeographic analysis and ancestral range reconstruction, offers a clearer view of the possible evolutionary history of the bent-toed geckos in the Himalayas.
4.1. Phylogeny of the Himalayan Bent-Toed Geckos
This study represents the most comprehensive analysis of Cyrtodactylus species across the region to date, sampling all the described species in the Himalayan range except C. dattanensis [47], for which we were unable to obtain DNA sequence data. A total of 46 homologous sequences were analyzed, including 22 additional new sequences to the most recent phylogeographic study of Himalayan geckos [7, 8]: 14 from the Central Himalayas, four from the eastern Himalayas south of the Brahmaputra River Basin, one from the eastern Himalayas north of the Brahmaputra, two from Indo-Burma south of the Brahmaputra, and one from east of the Irrawaddy River Basin were analyzed. The phylogenetic relationships among the geckos reconstructed in this study align with the findings of Agarwal et al. [7, 8], Wood et al. [12], and Grismer et al. [10, 11].
The phylogenetic analysis identified four major clades/groups of Himalayan geckos: the lawderanus group, the fasciolatus group, the peguensis group, and the khasiensis group, as proposed by Grismer et al. [10, 11]. The phylogeny reveals close relationships between the Cyrtodactylus lineages associated with the trans-Himalayan and western Himalayan species, consistent with previous studies by Grismer et al. [10, 11], and Wood et al. [12]. The nuclear and mitochondrial sequences of 14 specimens from the Central Himalaya represent four distinct taxonomical units in the Central Himalayan region. Species from the Doti and Chitwan regions of the Central Himalaya are grouped with the fasciolatus group. Notably, samples from the Doti areas show genetic differences of 14.9% from C. fasciolatus and 29.9% from C. lawderanus group from the Western Himalaya (Table S3). The specimens from Chitwan, approximately 815 km east of the known range of C. fasciolatus, also clustered in the fasciolatus group, showing a genetic difference of 29.8% from C. fasciolatus (Table S3). This confirms that the fasciolatus group extends its distribution from western Himalayas to the central region of the Central Himalayas. These findings are consistent with those of Grismer et al. [13], who predicted that species from the east of C. fasciolatus range adjacent to Nepal belong to the fasciolatus group, given their proximity to Nepal. Annual temperature, precipitation, and elevation may be associated with the distribution of the Himalayan geckos. The ANOVA analysis revealed significant associations with elevation (F = 3.382, p < 0.05) and annual temperature (F = 4.37, p < 0.05), but not with precipitation (F = 1.14, p > 0.05; Figure S3, Tables S4–S8). Precipitation is similar across the range of the four groups, averaging 2131 ± 1160 mm per year, indicating a preference for tropical humid climates [48]. Elevation and annual temperature are likely to influence their distribution due to their poikilothermic nature and preference for humid environments. The range of geckos within elevation and annual temperature is clade/group specific. The lawderanus group has significantly different means (1404 m, 17.0°C) compared to the fasciolatus (849 m, 20.9°C), khasiensis (616 m, 22.2°C), and peguensis (547 m, 22.0°C) groups, whereas the khasiensis, fasciolatus, and peguensis groups do not differ significantly from each other in elevation and annual temperature, respectively (Figure S3).
The genus Cyrtodactylus is the most diverse group of reptiles in the surrounding Himalayan region, with over 21 species described from southern Nepal, India, and Pakistan [7, 8, 10–12, 15, 49–52]. These geckos demonstrate remarkable dispersal capabilities and robust genetic adaptability, influencing the overall phylogenetic structure and diversity within the genus [10, 11, 13]. Their diversity is particularly remarkable in areas west of the Brahmaputra River to east of the Koshi River and west of the Irrawaddy River basin, which lie within both the Himalayan Biodiversity Hotspot and parts of the Indo-Burma Biodiversity Hotspot [7, 53–57]. While several research has been conducted in the eastern Himalayan region, leading to the remarkable discoveries of Cyrtodactylus in the last decade [14, 15, 58–61], large parts of the Central and Western Himalayas remain underexplored [54], which shed a gap for further study.
4.2. Biogeography of the Himalayan Bent-Toed Geckos
Our analysis supports the constant rate of speciation model with rapid increase of coalescent events in the recent population. Results from the LTT plot, BEAST analysis, and ancestral range reconstruction of the geckos in the Himalayas suggest a constant mode of genetic diversification within the specific geography of the Himalayan region, revealing a complex evolutionary pattern of diversification. The genus Cyrtodactylus, along with other gekkonid genera, is thought to have originated in what has been termed the “proto-Himalayan” region, or in the vicinity of the present-day Tibetan Plateau, around 62–52 mya [7, 12]. These early divergences are thought to be associated with the continuous compression of the Neotethyan island arcs located between the Indian subcontinent and Eurasia [18, 19].
During the early Paleogene, approximately 50.92 mya (53.96–47.85 mya), tectonic activity [2] combined with global climate changes, including the Paleocene–Eocene Thermal Maximum (PETM), had significant impacts [62, 63]. These changes created new habitats and ecological niches, driving vicariance and speciation, leading to the separation of western Himalayan species west of the Ganges from trans Himalayan species [64].
The inferred biogeographic events indicate with high probability and high confidence that species from the western Himalaya dispersed all over the Himalayan range to the west of Irrawaddy River basin of Myanmar. The dispersal events were continuous, rather than isolated, and occurred between 49.66 and 15.06 mya. We hypothesize the gradual process of multiple vicariance events followed by dispersal separated western species to species west of Irrawaddy River basin around 49.66 mya [65, 66]. Similarly, geckos from west of Ganges River basin separated from western and Central Himalayan species around 15 mya. This may have been due to continuous aridification between 20 and 10 mya, which created open arid areas and led to a lack of suitable microhabitats, water resources, and food. These harsh conditions may have forced them to migrate east of the Ganges, reducing gene flow and thus accelerating speciation [6, 35, 57, 67].
Comparatively, the highest level of diversification occurred in the eastern Himalaya, followed by the Central and Western Himalaya [7, 12] (Figure 4). Although species in the western Himalaya are unrepresented, ancestral range reconstruction analyses indicate west–east diversification of Cyrtodactylus. The four groups of Himalayan Cyrtodactylus got separated in the late Eocene, likely due to vicariance caused by the early uplift of the Himalayas between 45 and 35 mya [6]. Other major dispersal events occurred during two key geological periods: the Late Eocene-Early Oligocene and the Miocene. These events resulted in the separation of species mainly due to vicariance caused by persistent arid conditions between 20 and 10 mya, shifting monsoon patterns, and river formation due to orographic and plateau heating, which facilitated allopatric speciation between areas [5, 7, 12, 45, 57]. The diversity of Cyrtodactylus in the complex mountain landscape of Central and Western Himalaya remains underexplored and vastly underestimated [7]. Therefore, additional sampling from Central and Western Himalayas may reveal many undescribed species and better elucidate the phylogeography of the genus Cyrtodactylus.
5. Conclusions
Our analyses, covering geographical areas across the Himalaya and Indo-Burma region, support the use of Cyrtodactylus taxa as a model for understanding Himalayan biogeography and the impact of geological and climatic history on species diversity and dispersal. The constant rate of speciation in the Himalayan species of Cyrtodactylus suggests that no single factor plays a role in its diversification. Our results establish an intricate link between the orogenesis of the Himalaya, which creates landscape heterogeneity, global climate change, which leads to both hostile and favorable environments, persistent arid conditions, shifting monsoon patterns, and river formation. These factors have facilitated allopatric speciation of species in the geography studied. This study also supports a west–east diversification of Cyrtodactylus, despite constant genetic exchange within its range as in previous studies. The vast, favorable areas of the Himalayan region for the geckos remain largely unexplored due to their high genetic plasticity. Further sampling efforts are expected to uncover numerous additional undescribed species. Therefore, obtaining baseline information on the diversity and distribution across various taxonomic groups in this region is crucial for making informed decisions for effective biodiversity conservation.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Asmit Subba and Laxman Khanal designed the study, performed fieldwork, and laboratory analyses. Asmit Subba and Laxman Khanal analyzed the data and prepared the manuscript. Arjun Thapa revised the manuscript. All authors agreed with the final version of the manuscript.
Funding
This research was partly supported by the Second Tibetan Plateau Scientific Expedition and Research Program (STEP, 2019QZKK0501), China.
Acknowledgments
Laxman Khanal is grateful to the President’s International Fellowship Initiative (PIFI) program of the Chinese Academy of Sciences and Jiang Xue-Long from the Key Laboratory of Genetic Evolution and Animal Models, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China. Arjun Thapa is thankful to the Chinese Academy of Sciences for the PIFI Postdoctoral Fellowship. We thank the Department of Forest and Soil Conservation, Government of Nepal (DoFSC), for granting permission (Permission Number: 846/080/81) to conduct the research. Our thanks extend to Hammvai Lama, Ganesh Tamang, Laxmi Prasad Upadhya, Roshan Dulal, and Ashok Hamkhim Rai for their assistance during the fieldwork. Additionally, we appreciate the help of Sapana Ulak and Ankit Kumar Singh in laboratory analysis.
Supporting Information
Additional supporting information can be found online in the Supporting Information section.
Open Research
Data Availability Statement
The DNA sequence data used to support the findings of this study are available from the corresponding author upon request.




