Comprehensive Analysis of FBXO43 Expression and Its Prognostic and Therapeutic Implications in Osteosarcoma
Abstract
Background: Osteosarcoma (OS) is the most common malignant bone tumor in children and adolescents. FBXO43, a member of the F-box protein family, has been identified as a crucial prognostic factor in several cancers. However, its role in osteosarcoma remains largely unexplored.
Methods: This study investigated the expression and potential role of FBXO43 across the TCGA pan-cancer cohort, with a particular focus on sarcoma (SARC). We analyzed the relationship between FBXO43 expression and various cancer-related pathways, immune infiltration, and genomic features, including TP53 mutations. We also conducted immunotherapy predictive analyses using TIDE and Submap algorithms. To validate our findings, we performed a series of in vitro experiments, including cell invasion and wound healing assays.
Results: FBXO43 was found to be significantly overexpressed in various cancers and was particularly associated with pathways such as E2F_TARGETS and G2M_CHECKPOINT in SARC samples. Differential FBXO43 expression was linked to distinct immune infiltration patterns and pathway enrichments. Notably, high FBXO43 expression in osteosarcoma was associated with a higher TP53 mutation rate. Predictive analyses indicated that patients with low FBXO43 expression had better immunotherapy responses, suggesting its potential as a diagnostic and prognostic biomarker. These findings were corroborated by our in vitro functional assays.
Conclusion: Our comprehensive analysis reveals that FBXO43 plays a critical role in the progression of osteosarcoma, impacting both immune infiltration and genomic stability. FBXO43 expression levels may serve as valuable biomarkers for predicting immunotherapy responses in osteosarcoma patients. Future studies should aim to elucidate the molecular mechanisms driving FBXO43’s role and validating these findings in clinical settings to develop targeted therapeutic strategies.
1. Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor, predominantly affecting children and adolescents [1]. It arises from primitive bone-forming mesenchymal cells and is characterized by the production of osteoid or immature bone by the malignant cells [2]. Despite advancements in surgical techniques and chemotherapy, the prognosis for osteosarcoma remains poor, particularly in cases of metastatic or recurrent disease [3]. The five-year survival rate for localized osteosarcoma is approximately 60%–70%, but it drops sharply to 20%–30% in cases of metastasis [4]. Understanding the molecular mechanisms underlying osteosarcoma development and progression is crucial for identifying new therapeutic targets and improving patient outcomes. Recent research has shed light on the roles of various genetic and epigenetic factors in osteosarcoma pathogenesis, offering new opportunities for innovative treatment strategies [5].
FBXO43, also known as F-box only protein 43, belongs to the F-box protein family characterized by an F-box motif facilitating protein–protein interactions [6, 7]. F-box proteins play a pivotal role in the ubiquitin–proteasome pathway by targeting specific proteins for degradation, thereby regulating critical cellular processes such as cell cycle progression, apoptosis, and signal transduction [8, 9]. Recent studies suggest that FBXO43 may be implicated in cancer development and progression. Dysregulated expression of FBXO43 has been noted across various cancer types, where it is implicated in tumorigenesis by modulating key signaling pathways involved in cell proliferation and survival [10, 11]. For instance, in breast cancer (BC), reduced FBXO43 expression inhibits tumor growth by attenuating its interaction with PCNA, highlighting FBXO43 as a potential oncogenic driver and therapeutic target in BC [12]. Moreover, FBXO43 has been identified as a significant gene associated with osteonecrosis of the femoral head, linked closely to immune infiltration imbalances observed in this condition [13]. To date, while FBXO43 has been shown to play a significant role in osteonecrosis of the femoral head, its role in osteosarcoma remains unclear and warrants further investigation.
In our study, we selected FBXO43 based on a thorough review of the literature, recognizing its potential significance in cancer biology. We initiated our investigation with in vitro experiments, including colony formation assay and transwell migration assay, to establish foundational insights into FBXO43’s functional role in osteosarcoma. Subsequently, leveraging bioinformatics analyses, we explored its expression and potential oncogenic functions across various cancers, with a specific emphasis on sarcomas, particularly osteosarcoma. Furthermore, predictive analyses for immunotherapy underscored the impact of FBXO43 expression on treatment responses in osteosarcoma patients. These integrated approaches aimed to validate and broaden our understanding of FBXO43’s role in cancer progression and therapeutic response.
2. Methods
2.1. Cell Culture
The human osteoblastic cell line hFOB, along with the human osteosarcoma cell lines MG63 and U2OS, were used. The hFOB cells were cultured at 37°C in a 5% CO2 atmosphere using phenol red-free Ham’s F12/DMEM (1:1) medium supplemented with 10% fetal bovine serum (FBS) and 0.3 mg/mL G418. Both MG63 and U2OS cells were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% FBS. All cell lines were routinely passaged upon reaching approximately 80% confluence, ensuring exponential growth during experiments.
2.2. Cell Transfection and Lentiviral Packaging
For the knockdown of FBXO43, shRNA sequences targeting FBXO43 were cloned into the pLKO.1 plasmid backbone (target sequence: 5′-CAAGTTATCAACTTAGAAA-3′). The resulting sh-FBXO43 constructs were cotransfected with the packaging plasmids pMD2.G and psPAX2 into HEK293T cells to produce lentiviral particles. This transfection employed a three-plasmid system. HEK293T cells were plated at a density of 2.5 × 106 cells per 10 cm dish one day before transfection. Transfection was performed using Lipofectamine 2000, according to the manufacturer’s instructions. In brief, for each dish, 10 μg of sh-FBXO43 plasmid, 7.5 μg of psPAX2, and 2.5 μg of pMD2.G were mixed with Opti-MEM I reduced serum medium and combined with Lipofectamine 2000. The DNA-Lipofectamine complex was added to the cells in a dropwise manner. After 48 h post-transfection, the supernatant containing lentiviral particles was collected, filtered through a 0.45 μm filter to remove cellular debris. The viral pellet was resuspended in a suitable medium for further applications in target cell transduction studies.
2.3. Cell Counting Kit-8 (CCK-8) Assay
For the CCK-8 assay, cells were seeded in 96-well plates at a density of 1000 cells per well. After 24 h to allow for cell attachment, the medium was replaced, and cells were treated as per the experimental design. Cell viability was assessed at 24, 48, and 72 h posttreatment. At each time point, 10 μL of CCK-8 solution was added to each well, and the plates were incubated for an additional 2 h at 37°C. Absorbance was measured at 450 nm using a microplate reader.
2.4. Colony Formation Assay
For the colony formation assay, cells were plated in 6-well plates at a low density (500 cells per well) to allow individual colonies to form. After 24 h, the cells were treated according to the experimental conditions and allowed to grow for 2 weeks. The medium was refreshed every 3 days. At the end of the incubation period, colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Colonies consisting of more than 50 cells were counted.
2.5. Transwell Migration Assay
To evaluate the migratory capacity of cells following FBXO43 knockdown, a Transwell migration assay was performed using 24-well plates with inserts containing 8-μm pore size membranes. Cells were trypsinized and resuspended in serum-free medium after 48 h post-transduction. A total of 1 × 105 cells in 200 μL of serum-free medium were seeded into the upper chamber of each insert, while 600 μL of medium containing 10% FBS, which served as a chemoattractant, was added to the lower chamber. The cells were allowed to migrate for 24 h at 37°C in a 5% CO2 atmosphere. Nonmigrated cells on the upper surface of the membrane were gently removed using a cotton swab. Migrated cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal violet for 20 min, and rinsed with water. Migrated cells were quantified by counting five random fields per membrane under a light microscope at 100x magnification.
2.6. Data Acquisition and Analysis of FBXO43 in Pan-Cancer and SARC
First, we obtained RNA sequencing data and corresponding clinical information from The Cancer Genome Atlas (TCGA) database. This dataset encompassed various cancer types, enabling comprehensive pan-cancer analysis. Using R software and the “edgeR” package, we compared FBXO43 expression levels between cancerous and normal tissues across multiple cancer types. For functional enrichment analysis, we employed the Hallmark gene sets from the Molecular Signatures Database (MSigDB). Gene Set Enrichment Analysis (GSEA) was performed to identify pathways and biological processes significantly associated with FBXO43 expression. Finally, to further explore the specific role of FBXO43 in sarcoma (SARC), we conducted Gene Ontology (GO) analysis to determine the biological processes, cellular components, and molecular functions enriched in the FBXO43-associated gene set within SARC samples.
2.7. Prognostic Analysis of FBXO43 in Pan-Cancer and SARC
Prognostic data, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI), were sourced from the UCSC Xena database (https://xenabrowser.net/datapages/). We used univariate Cox regression to evaluate the prognostic significance of continuous FBXO43 expression levels across various cancer types. In addition, to specifically assess the prognostic value of FBXO43 in sarcoma (SARC), we conducted Kaplan–Meier survival analysis within the SARC cohort. This analysis employed dichotomized FBXO43 expression levels, with cutoff points determined using the “surv-cutpoint” function from the “survminer” R package. The log-rank p value, hazard ratio (HR), and 95% confidence interval (95% CI) were calculated to evaluate the impact of FBXO43 expression on patient survival outcomes, ensuring a comprehensive and robust assessment of its prognostic value.
2.8. Single-Cell Analysis of FBXO43 Using the Tumor Immune Single-cell Hub (TISCH) Database
We conducted single-cell analysis of FBXO43 using TISCH database to explore the expression and cellular context of FBXO43 in osteosarcoma. Initially, we annotated cell types within the osteosarcoma dataset GSE162454. Subsequently, we investigated the distribution of FBXO43 expression across various cell types within this dataset. Additionally, we performed an analysis of transcriptional regulators and intercellular interactions to elucidate the regulatory mechanisms involving FBXO43 in osteosarcoma. This approach allowed us to gain insights into the specific cellular environments where FBXO43 may exert its functional roles, providing a comprehensive view of its potential impact within the tumor microenvironment.
2.9. Analysis of FBXO43 in Relation to Tumor Mutation Burden (TMB), Microsatellite Instability (MSI), Purity, and Mutational Landscape
Considering the strong correlation between TMB and the effectiveness of PD-1/PD-L1 inhibitors, as well as the critical roles of MSI and tumor purity in cancer progression, we investigated FBXO43 expression levels in relation to TMB, MSI, and tumor purity across various cancers. Furthermore, we analyzed the mutational landscape of FBXO43 across different cancer types, focusing on both the frequency and types of mutations. In the SARC cohort specifically, we compared the mutational profiles of patients with high versus low FBXO43 expression levels. This analysis included evaluating differences in mutation rates and mutation types, offering valuable insights into the genetic variations linked to FBXO43 and their potential implications for cancer development and therapeutic strategies.
2.10. Functional Analysis of FBXO43 in Osteosarcoma Cohort
Using the osteosarcoma dataset TARGET-OS, we divided patients into high and low FBXO43 expression groups. We then performed GO and KEGG pathway analyses to understand the functional differences between these groups. For GO analysis, we used the ClueGO plugin in Cytoscape. ClueGO allows for the visualization of GO enrichment results in the form of network diagrams, facilitating the interpretation of complex biological data. The enriched GO terms were identified and visualized to highlight the biological processes, molecular functions, and cellular components associated with FBXO43 expression. KEGG pathway analysis was also conducted to classify and understand the biological pathways involved. The results of the KEGG analysis were categorized to provide a clearer overview of the pathways significantly associated with high and low FBXO43 expression levels.
2.11. Immune Infiltration and Immunotherapy Response Analysis
To assess the differences in the immune microenvironment between high and low FBXO43 expression groups, we conducted immune infiltration analysis. This involved evaluating StromalScore, ImmuneScore, and ESTIMATEScore differences between the two groups. In addition, we analyzed the expression of common immune checkpoint-related genes to understand the immunogenic landscape influenced by FBXO43. For predicting immunotherapy response, while PD-L1 expression is commonly used, we chose the tumor immune dysfunction and exclusion (TIDE) and Submap algorithms due to their ability to provide a comprehensive assessment beyond PD-L1. The TIDE algorithm predicts immune escape mechanisms by assessing T-cell dysfunction and exclusion, factors critical in osteosarcoma’s complex immune interactions [14]. Submap compared our gene expression profiles with known immunotherapy responders and nonresponders, validating potential outcomes across various cancers, including osteosarcoma [15]. These methods were selected to complement traditional biomarkers and provide a deeper understanding of immunotherapy potential in osteosarcoma, addressing the limitations of PD-L1 in capturing its full complexity.
3. Results
3.1. Effects of FBXO43 Knockdown on Osteosarcoma Cell Lines
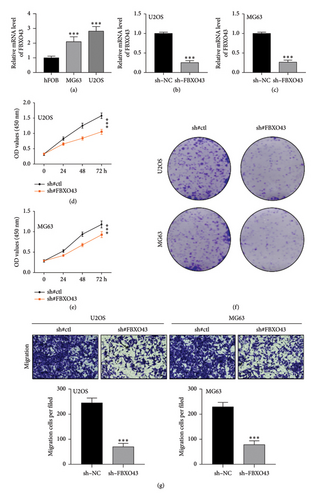
Figure 1(a) illustrates that FBXO43 is expressed in the human osteoblastic cell line hFOB and the human osteosarcoma cell lines MG63 and U2OS at varying levels. Following FBXO43 knockdown, Figures 1(b) and 1(c) show a significant reduction in its expression in U2OS and MG63 cell lines, confirming the knockdown’s efficiency. The CCK-8 assay results (Figures 1(d) and 1(e)) reveal that cell viability significantly decreased at 24, 48, and 72 h posttreatment in both cell lines after FBXO43 knockdown. In addition, Figure 1(f) shows that colony formation was markedly reduced following FBXO43 knockdown. The Transwell migration assay results (Figure 1(g)) indicate that the migratory capacity of U2OS and MG63 cells was significantly impaired postknockdown, highlighting FBXO43’s role in tumor cell migration.
3.2. FBXO43 Expression and Pathway Enrichment Analysis in Pan-Cancer and SARC
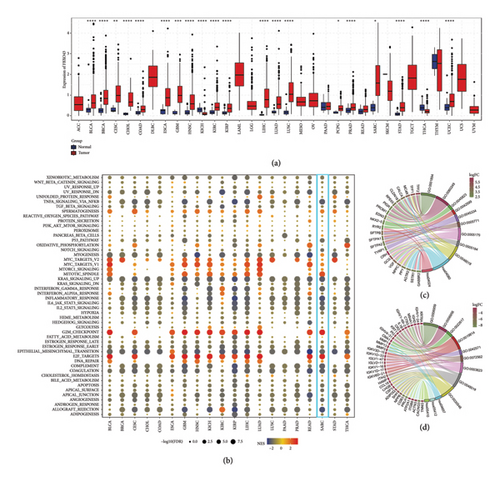
Figure 2(a) illustrates the expression levels of FBXO43 across a range of cancer types, demonstrating that FBXO43 is markedly overexpressed in most cancers, including SARC. This widespread overexpression implies a potential oncogenic function for FBXO43. In Figure 2(b), pathway enrichment analysis of FBXO43 in a pan-cancer context reveals its association with several key pathways, particularly in SARC, where it is linked to critical processes such as E2F_TARGETS and G2M_CHECKPOINT, both of which play pivotal roles in cell cycle regulation and proliferation. Figures 2(c) and 2(d) present GO and KEGG pathway analyses comparing high and low FBXO43 expression groups in SARC. These analyses uncover significant differences in biological processes, molecular functions, and cellular components, shedding light on the potential involvement of FBXO43 in the development and progression of sarcoma.
3.3. Prognostic Role of FBXO43 Across Various Cancers
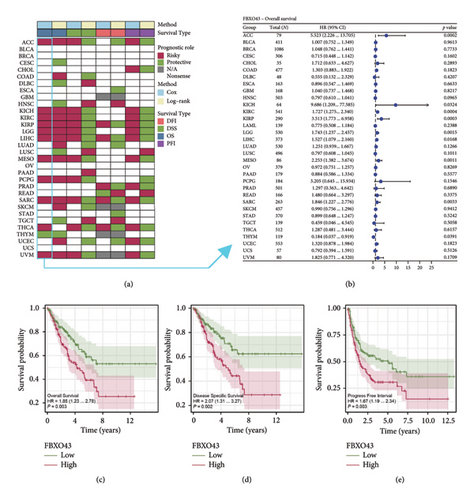
Our prognostic analysis of FBXO43 across multiple cancers revealed that FBXO43 often acts as a risk factor (Figure 3(a)). The forest plot in Figure 3(b), generated using univariate Cox regression analysis, further illustrates the prognostic impact of FBXO43 across different cancer types, highlighting its association with poor outcomes in most cases. Furthermore, in the SARC cohort, KM survival analyses were conducted, focusing on OS, DSS, and PFI. As shown in Figures 3(c) and 3(d), the results indicate that high FBXO43 expression is significantly associated with worse outcomes in all these survival metrics, underscoring its potential as a prognostic marker in sarcoma.
3.4. FBXO43 Expression and Interaction in Osteosarcoma at the Single-Cell Level
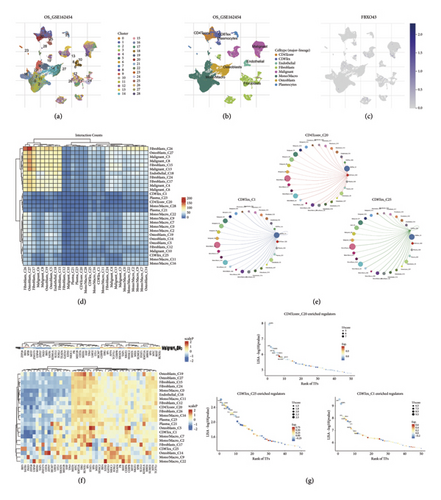
Using the TISCH2 database, we visualized the cell type annotations and expression patterns within the osteosarcoma dataset OS_GSE162545 (Figures 4(a) and 4(b)). We then explored the distribution of FBXO43 expression across these annotated cell types, revealing distinct expression profiles (Figure 4(c)). Further analysis of cell–cell interactions highlighted significant communication networks among different cell types (Figures 4(d) and 4(e)), providing insights into the complex intercellular signaling within the tumor microenvironment. In addition, transcription factor enrichment analysis identified several key transcription factors involved in the regulatory networks (Figures 4(f) and 4(g)), offering a deeper understanding of the transcriptional regulation in osteosarcoma.
3.5. Correlation of FBXO43 With TMB, MSI, Purity, and Mutational Landscape
We examined the correlation between FBXO43 expression and TMB, MSI, and tumor purity across various cancers. Our analysis revealed that FBXO43 expression is positively correlated with TMB, MSI, and tumor purity in most cancers, with a particularly strong correlation observed in SARC (Figures 5(a), 5(b), and 5(c)). Figure 5(d) illustrates the mutational landscape of FBXO43 across different cancer types, highlighting the frequency and types of mutations. In the SARC cohort, we compared the mutational profiles between patients with high and low FBXO43 expression levels. The results show distinct differences in mutation patterns between the two groups, suggesting a potential link between FBXO43 expression and the mutational characteristics of sarcoma (Figure 5(e)).
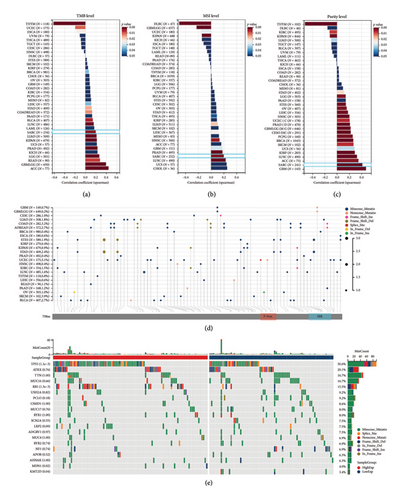
3.6. Functional Analysis of High and Low FBXO43 Expressions in Osteosarcoma
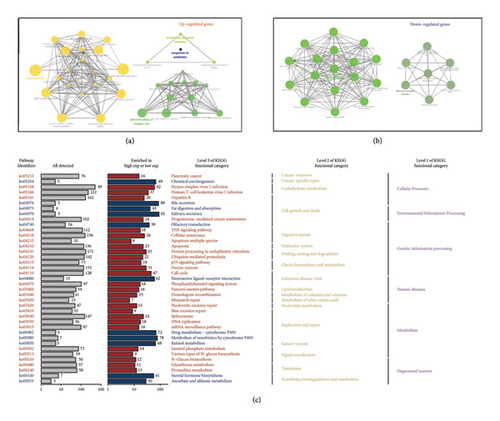
Based on the osteosarcoma dataset TARGET-OS, we divided patients into high and low FBXO43 expression groups. The GO analysis results for the two groups are shown in Figures 6(a) and 6(b). In the high FBXO43 expression group, enriched pathways included “response to antibiotic,” “detoxification of copper ion,” and “nucleosome assembly.” Conversely, in the low FBXO43 expression group, enriched pathways included “type I interferon receptor binding” and “olfactory receptor activity.” The KEGG pathway analysis revealed significant differences between the high and low FBXO43 expression groups. In the high-expression group, pathways such as “TNF signaling pathway,” “apoptosis,” and “p53 signaling pathway” were enriched. In contrast, the low-expression group showed enrichment in pathways related to “drug metabolism” and “retinol metabolism” (Figure 6(c)). These results indicate distinct functional and biological differences driven by FBXO43 expression levels in osteosarcoma, highlighting its potential impact on tumor behavior and patient prognosis.
3.7. Immune Landscape and Immunotherapy Efficacy Associated With FBXO43 in Osteosarcoma
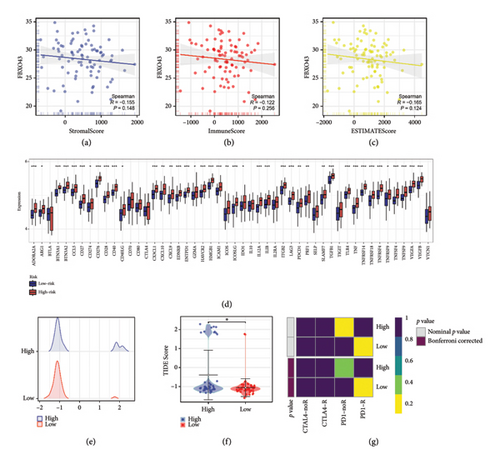
Our immune infiltration analysis revealed that FBXO43 expression in osteosarcoma is negatively correlated with immune infiltration levels, as indicated by the StromalScore, ImmuneScore, and ESTIMATEScore (Figures 7(a), 7(b), and 7(c)). In addition, there were significant differences in the expression levels of immune checkpoint-related genes between the high and low FBXO43 expression groups (Figure 7(d)). Furthermore, Figure 7(e) displays the distribution of TIDE scores among osteosarcoma patients. The TIDE analysis indicated that patients in the low FBXO43 expression group had lower TIDE scores, suggesting a potentially better response to immunotherapy (Figure 7(f)). This finding is corroborated by the Submap analysis results, which also demonstrated that the low-risk group patients are more likely to respond favorably to immunotherapy (Figure 7(g)).
4. Discussion
Osteosarcoma, the most common malignant bone tumor in children and adolescents, is associated with a high mortality rate [16]. With the rapid advancement of sequencing technologies, bioinformatics has emerged as a powerful tool in cancer research, enabling the analysis of complex biological data and uncovering novel insights into cancer mechanisms [17]. The application of bioinformatics in osteosarcoma research has significantly advanced our understanding of the molecular pathways driving this aggressive malignancy. For instance, integrative genomic analyses have identified key genetic alterations and pathways involved in osteosarcoma pathogenesis, offering insights into potential personalized treatment strategies [18]. Moreover, bioinformatics approaches have facilitated the detailed analysis of gene expression profiles and regulatory networks, enhancing our ability to predict patient prognosis and response to therapy [19].
In this study, we employed a distinct experimental approach, initiating with in vitro assays prior to performing bioinformatics analyses. This decision was informed by a comprehensive review of the literature, which underscored the pivotal role of FBXO43 in various cancers. Notably, FBXO43 has been implicated in essential cellular processes such as cell cycle regulation and tumor proliferation. For example, FBXO43 is a critical prognostic factor in multiple cancers and plays a significant role in osteonecrosis of the femoral head [20, 21]. However, its intrinsic functions and molecular mechanisms in osteosarcoma remain largely unknown. Given these insights, we hypothesized that FBXO43 could play a similarly critical role in osteosarcoma, a malignancy in which its function remains largely unexplored. To test this hypothesis, we first investigated the functional significance of FBXO43 in osteosarcoma cell lines through in vitro assays, including evaluations of cell viability, migration, and invasion. These initial experiments were instrumental in establishing FBXO43’s relevance in osteosarcoma biology, providing a foundation for subsequent bioinformatics exploration.
Subsequently, we were the first to investigate FBXO43 expression across the TCGA pan-cancer cohort, identifying its association with multiple cancer-related pathways. Importantly, FBXO43 appears to play a pivotal role in the development and progression of osteosarcoma. Patients with varying levels of FBXO43 expression exhibited notable differences in immune infiltration levels. In addition, significant differences were observed in the enriched pathways between high and low FBXO43 expression groups. Crucially, multiple predictive analyses for immunotherapy response indicated that patients with low FBXO43 expression may have better immunotherapy outcomes, highlighting the potential diagnostic and prognostic value of FBXO43. These findings point toward its possible integration into future therapeutic strategies, especially in the context of immunotherapy.
In SARC samples, we found that FBXO43 expression is closely associated with the E2F_TARGETS and G2M_CHECKPOINT pathways, both of which play crucial roles in cancer progression. On the one hand, E2F transcriptional activity is meticulously controlled throughout the cell cycle through various mechanisms. Disruptions in one or more key components of the cyclin-dependent kinase (CDK)-RB-E2F axis are present in nearly all cancers, resulting in elevated oncogenic E2F activity and subsequent uncontrolled cellular proliferation [22]. A study has indicated that miR-145-5p may inhibit the progression of osteosarcoma, suggesting its potential as a valuable biomarker for diagnosis and a therapeutic target for treatment [23]. On the other hand, the G2M checkpoint in cell cycle regulation facilitates the transition from the G2 phase to the M phase through cyclin B-cdc2 (CDK1) complexes and the control of this transition by CDK1 plays a crucial role in tumorigenesis [24, 25]. In addition, a study has demonstrated that the G2M pathway score can serve as a valuable tool for identifying patients with ER-positive/HER2-negative breast cancer who are at a higher risk of metastasis and have poorer survival outcomes [26]. These findings suggest that FBXO43 may be involved in the aforementioned pathways, influencing the development and progression of SARC, which opens new avenues for further in-depth research.
In our comprehensive analysis of genomic characteristics, we observed a notable trend: osteosarcoma patients with elevated FBXO43 expression exhibited a higher frequency of TP53 mutations. This finding suggests a potential link between elevated FBXO43 levels and genomic instability, underscoring its significance of FBXO43 in the pathogenesis of osteosarcoma. TP53, often referred to as the “guardian of the genome,” is crucial for regulating cell cycle progression and maintaining genomic integrity [27]. Mutations in TP53 are well known to drive uncontrolled cell proliferation and tumor progression [28]. The increased TP53 mutation rate in patients with elevated FBXO43 expression may indicate a synergistic interaction, fostering tumor aggressiveness and resistance to conventional therapies. These findings suggest that FBXO43 could serve as a biomarker for identifying high-risk osteosarcoma patients and emphasize the need for further studies to clarify the molecular mechanisms underlying this association. Understanding the interplay between FBXO43 and TP53 mutations could inform the development of targeted therapies aimed at improving outcomes for patients with osteosarcoma.
In our analysis, osteosarcoma samples with low FBXO43 expression exhibited a more favorable response to immunotherapy, as demonstrated by both the TIDE and Submap algorithms. This finding suggests that FBXO43 expression levels may be inversely correlated with the efficacy of immunotherapeutic interventions. The TIDE algorithm, which predicts immune escape potential, and the Submap algorithm, which compares gene expression profiles to known responders and nonresponders to immunotherapy, both indicated a better therapeutic response in patients with lower FBXO43 expression. These findings imply that higher FBXO43 expression could be linked to a more immunosuppressive tumor microenvironment, potentially reducing the effectiveness of immune checkpoint inhibitors and other immunotherapies. Conversely, lower FBXO43 expression may promote a more robust immune response against tumor cells, enhancing the efficacy of immunotherapy.
These findings underscore the potential of FBXO43 as a predictive biomarker for immunotherapy responsiveness in osteosarcoma. However, several limitations of our study should be acknowledged. First, the retrospective nature of the data analysis may introduce selection bias, and thus, our findings require validation in prospective clinical trials. Second, while the TIDE and Submap algorithms offer valuable insights into immunotherapy response, their predictions should be interpreted with caution and confirmed by clinical outcomes. Lastly, the heterogeneity of osteosarcoma and the complexity of the tumor microenvironment may limit the generalizability of our results. Future studies should incorporate larger, more diverse patient cohorts and utilize multiomics approaches to achieve a more comprehensive understanding of FBXO43’s role in osteosarcoma.
Ethics Statement
The authors have nothing to report.
Conflicts of Interest
The authors declare that they have no competing interests.
Author Contributions
Jiang Jin, Kang Yao, and Zhao Tingxiao supervised the project design and offered comprehensive guidance. Yao Longtao handled the data collection from the TCGA database. Li Yanlei and Tian Jinlong performed the data analysis and prepared the initial manuscript. Zhu Xiaojun and Han Liang thoroughly reviewed and revised the manuscript, ensuring significant intellectual content. Jiang Jin, Kang Yao, and Zhao Tingxiao contributed equally to this work.
Funding
This study was funded by Zhejiang Provincial Health Bureau Science Foundation of China, No. 2024KY647 Found and Zhejiang Provincial Natural Science Found LQ24H160045.
Acknowledgment
We would like to acknowledge that we used the ChatGPT-4 model (https://www.openai.com/chatgpt) for text polishing of the entire manuscript during the final revisions. Also, a preprint has previously been published [29].
Open Research
Data Availability Statement
The data used in this study including TCGA-SARC and TARGET-OS datasets are available from The Cancer Genome Atlas (TCGA) database. Additional information is available from the corresponding author upon reasonable request.




