Molecular Characterization and Phylogenetic Analysis of Velogenic NDV Genotype VII Isolated From an Outbreak in Genotype II-Vaccinated Poultry Flocks of the Southern Region of Pakistan
Abstract
Despite the administration of multiple doses of the Newcastle disease virus (NDV) vaccine, a high mortality rate (20%–40%) due to NDV was observed in the southern region of Pakistan. In the current study, clinical samples were obtained from affected areas with NDV infection. The clinical examination revealed characteristics signs and symptoms of NDV and 40% mortality. Furthermore, necropsy revealed characteristic lesions commonly associated with NDV infection. NDV antibodies were detected by ELISA in 27% of breeders, 45% of layers, and 62% of broiler flocks. The RT-PCR confirmed the infection in 15% of breeders, 17% of layers, and 21% of broilers. Phylogenetic analysis of the Fusion gene revealed that the outbreaks in vaccinated flocks of three districts were hit by NDV Genotype VII and in one district by Genotype II. The virulence factors of local isolates were determined based on amino acid analysis of the Fusion protein cleavage motif from 112–117 sites. The virulence factors (amino acids 112–117) of local isolates were compared with the virulent factors of the Fusion protein of the reported strains. All the new isolates from the three districts were found to contain polybasic amino acid 112 RRQRR↓F117 with F at the 117th position in the Fusion protein cleavage motif, which is a characteristic of the velogenic type of NDV. Isolates from one of the districts contained the monobasic amino acid 112 G, R, Q, G, R↓L 117 at the cleavage site, which is considered as an avirulent NDV type. Some substitution mutations in the amino acids of Fusion protein were observed among the new isolates, where the amino acid serine (S) was replaced by proline (P) at position 105, and serine/thymine (S/T) was replaced by alanine (A) at position 107. The data partially suggest that the NDV outbreak in the southern region of Pakistan with variant genetic group Genotype VII corresponds to a velogenic pathotype, and the mutations may be responsible for enhanced fatal consequences in affected flocks.
1. Introduction
The southern region of Pakistan, specifically Sindh (located at 25.8943° N, 68.5247° E) [1], plays a significant role in the transmission and spread of Newcastle disease virus (NDV) due to its geographical and ecological characteristics. This region, which includes a vast coastal area, lakes like Khanjar and Manchar, and dams, is a major hub for migratory birds and hosts a rich variety of wild fauna. The presence of diverse bird species, including cormorant ducks and geese, during the winter season, creates a favorable environment for the incubation, propagation, mutation, and transmission of disease particularly NDV, to commercial poultry flocks.
Previous studies reported the prevalence of velogenic NDV in wild migratory birds [2–4]. The wild bird’s comomorant is considered a potential source for the spread of NDV in South Asia [5]. The seasonal migration of wild birds contributes to the widespread of disease within the same or other species [6–9]. Highly virulent NDV strains are commonly found in all types of domestic poultry, from pigeons to ostriches, and in wild birds. Capturing wild birds and caging them with fancy birds in live wet markets in mixed bird populations provide a favorable environment for the emergence, transmission, and spread of NDV viruses [10]. NDV infects many avian species of birds and mammals [11] with a wide range of mortality [12] and causes severe economic impacts [13]. It causes asymptomatic to systematic infection with severe clinical manifestations that depend on the virulence of the NDV [14].
Newcastle disease is an economically important neurological and enteric disease in poultry. The causative agent of NDV is a negative sense single-stranded nonsegmented RNA virus. NDV belongs to the genus Avulavirus and the Paramyxoviridae family [15]. The NDV genome encodes six proteins, namely, nucleoprotein (NP), phosphoprotein (PP), matrix (M) protein, Fusion protein, hemagglutinin–neuraminidase (HN), and the RNA-dependent RNA polymerase (L) [16]. NDV strains are classified into a single serotype, but there is significant genetic and antigenic variation among different strains. This diversity has led to the classification of NDV into different genotypes and subgenotypes based on partial or complete sequences of the Fusion gene [17–19]. Based on genome size, avian paramyxovirus type-1 (APMV-1) is classified into two classes: Class I and Class II. Class I NDVs consist of apathogenic viruses that are usually found in wild birds and rarely found in poultry. Class II NDVs encompass low virulence (lentogenic), medium virulence (mesogenic), and highly virulent (velogenic) pathotypes [20]. These strains have been detected in various species of commercial, domestic and wild birds worldwide. Class I NDV has a single genotype, while Class II NDV has 20 distinct genotypes [21–24]. Different genetic groups are prevalent in different geographical regions of the world [25]. The clinical manifestation of NDV infection is variable, mild to severe, with a high range of mortality [26].
Different techniques such as ICPI, IVPI, and MDT are used to evaluate the pathogenesis of NDV [27]. The availability of gene sequencing has added a great understanding of NDV pathogenicity at the molecular level [15, 1621]. NDV strains are highly prone to mutations leading to the evolution of new variants or genotypes [28]. The NDV Fusion protein is considered a major determinant of NDV pathogenicity; cleavage of F0 into F1 and F2 makes the NDV infectious, enabling the fusion of the viral envelope with the host membrane [29]. A variable amino acid sequence is found at the Fusion protein cleavage site. The amino acid sequence of the Fusion cleavage site determines the ubiquitous cleavage of the Fusion protein by different proteases, present in multiple organs of the host. Strains containing the polybasic amino acid R/G/KR-Q/K-K/R-R↓F (RRQRR or RRQKR), at their cleavage site, can be cut by a wide range of proteases present in the host, allowing the virus to spread to multiple organs and become more infectious or highly virulent. However, avirulent strains have monobasic amino acids that can be cleaved by a limited range of proteases present in the host [29, 30]. The monobasic sequence 112GR/K-Q-G-R↓L117 can only be cleaved by trypsin-like enzymes present in the upper respiratory tract; hence, fatal infections are limited to the upper respiratory tract [31–35].
With the expansion of the poultry industry in Pakistan, NDV has become a major concern for poultry farmers, posing significant threats to the vaccinated flocks. This situation might be threatened by the rich wild fauna in the water habitat areas of the southern region. Previous reports indicate the circulation of different genotypes in commercial and backyard poultry in various parts of the Punjab province [36–38]. Owing to dense poultry sites, various studies have been conducted with particular emphasis on the molecular epidemiology of NDV in Punjab. However, this area of research has remained neglected to explore epidemiological markers for poultry in the southern part of the country. Such lapse needs to be filled as the NDV outbreak has commenced in vaccinated flocks with huge mortality. In the region where poultry flocks are vaccinated with live and killed vaccines with repeated shots, failure to protect against field challenge summons further molecular investigation. Therefore, isolation and identification of NDV from vaccinated flocks, gene sequencing, strain typing, and genetic grouping of field strains are of critical importance. The aim of this study was to isolate and identify NDV from vaccinated flocks, evaluate the pathogenicity of NDV isolates by analyzing the Fusion protein, and develop a phylogenic link between new isolates and different epidemiological or genetic groups, circulating in the world, by utilizing RT-PCR, sequencing, and phylogenetic analysis of NDV Fusion gene.
2. Materials and Method
2.1. Study Area
The study area for this investigation encompassed four key districts in the southern region of Pakistan, which are rich in water habitat, that is, near coastal regions (Karachi and Hyderabad), lakes (Thatta), and river (Dadu), with a predominance of commercial birds and dense population of migratory birds that seasonally reside near the targeted study areas.
2.2. Collection of Samples
A total of 877 serum and 370 tissue samples were collected from commercial poultry flocks including breeders, layers, and broilers of varying ages across the selected areas. Samples were collected under aseptic conditions from the lungs, intestines, and trachea of diseased, freshly dead, and euthanized birds. The adapted vaccination schedule in the investigated flock is given in Table 1. The vaccination schedule in the investigated flocks included multiple shots of live and inactivated NDV vaccines singly or in combination with other vaccines [39].
| Routine vaccination schedule of NDV usually practiced in different flocks | ||||||
|---|---|---|---|---|---|---|
| Sr. No. | No. of samples | Flock type | Age at sample collection | Age in days | Schedule of vaccination adapted in Sindh | |
| Tissue/swab samples | Blood samples | |||||
| 1 | 35 | 71 | Broiler | 7–30 D | 1 D | ND (hatchery) |
| 5 D | ND 1B | |||||
| 12 D | NDV Lasota | |||||
| 21 D | NDV clone | |||||
| 2 | 42 | 55 | Broiler | 7–30 D | 1 D | ND (Hatchery) |
| 5 D | ND 1B | |||||
| 15 D | NDV Lasota | |||||
| 24 D | NDV clone | |||||
| 3 | 64 | 108 | Broiler | 7–30 D | 1 D | ND (Hatchery) |
| 7 D | ND 1B | |||||
| 15 D | NDV Lasota | |||||
| 28 D | NDV clone | |||||
| 4 | 51 | 207 | Breeder | 10–80 W | 1D | NDV + IBV(Hatchery) |
| 7 D | NDV Lasota | |||||
| 14 D | IBH + NDV killed | |||||
| 28 D | ND + IBV live | |||||
| 35 D | ND + IBV killed | |||||
| 42 D | IBH + NDV killed | |||||
| 63 D | NDV Lasota | |||||
| 100 D | NDV Lasota | |||||
| 126 D | NDV + IBV | |||||
| 5 | 56 | 180 | Breeder | 10–80 W | 1D | NDV + IBV (hatchery) |
| 5 D | NDV Lasota | |||||
| 14 D | IBH + NDV killed | |||||
| 30 D | ND + IBV live | |||||
| 35 D | ND + IBV killed | |||||
| 42 D | IBH + NDV killed | |||||
| 60 D | NDV Lasota | |||||
| 80 D | NDV Lasota | |||||
| 133 D | NDV + IBV + EDS | |||||
| 6 | 28 | 63 | Layer | 8–110 W | 1 D | ND + IB (hatchery) |
| 7 D | NDV + IBV killed | |||||
| 28 D | IB + NDV live | |||||
| 35 D | ND + IBV killed | |||||
| 60 D | IB + NDV live | |||||
| 133 D | NDV + IB live | |||||
| 7 | 52 | 115 | Layer | 8–110 W | 1 D | ND + IB (hatchery) |
| 6 D | ND Lasota | |||||
| 7 D | NDV + IBV killed | |||||
| 28 D | IB + NDV live | |||||
| 35 D | ND + IBV killed | |||||
| 60 D | IB + NDV live | |||||
| 140 D | NDV + IB live | |||||
| 8 | 42 | 78 | Layer | 8–110 W | 1 D | ND + IB (hatchery) |
| 6 D | ND Lasota | |||||
| 7 D | NDV + IBV killed | |||||
| 28 D | IB + NDV live | |||||
| 35 D | ND + IBV killed | |||||
| 60 D | IB + NDV live | |||||
| 142 D | NDV + IB live | |||||
| Total | 370 | 877 | ||||
- Abbreviations: D, days; W, weeks.
2.3. Blood Sample Collection
Blood samples were collected from the wing veins of the affected birds using a syringe (3 mL). The wing was pulled outward until the wing vein became evident. Feathers that obscured the vein were plucked away. The area around the bleeding site was disinfected with 70% alcohol. The needle was inserted under the tendon and directed to the wing vein in the direction of the blood flow avoiding any deep insertion of the needle. Once the tip of the needle was in the vein, blood was allowed to flow into the syringe by gently pulling the plunger. The collected blood was immediately transferred to vacutainer tubes and kept at 2°C–8°C for 12 h for separation of serum. The separated serum was then collected and heat-inactivated by incubating at 56°C for 1 h and stored at 2°C–8°C for immediate use and at −80°C for long-term storage.
2.4. Necroscopy and Collection of Tissue Sample
Primarily, birds were screened for infection based on clinical signs (coughing sneezing, lacrimation, swelling of the eye, and wattles), morbidity, and mortality. The selected severely morbid birds were isolated by housing separately and then euthanized by slaughtering. The slaughtered birds were laid on their backs, examined externally, and then wet ventrally with soapy water. The skin was cut and peeled back from the midline of the chest to expose the muscle. The midline of the chest was sheared with scissors, and the birds were opened with forceps; specific lesions were identified in targeted organs, for example, the intestine, proventriculus, trachea, and lungs. Tissue samples were collected from the targeted organs.
2.5. Virus Isolation From Tissue Samples
2.5.1. Virus Isolation by Embryonated Eggs
The collected tissue samples were placed in the transport medium containing 1 mL of 50% glycerol, 50% phosphate-buffered saline, streptomycin (200 mg/L), penicillin (2 × 106 U/L), and amphotericin B (250 mg/L) and immediately transferred to the lab. Tissue homogenate was prepared by mincing 0.5 g tissue per 1.5 mL PBS, and virus suspension was prepared by centrifugation at 10,000 × g for 5 min and then passing the supernatant through a 0.45-μm membrane filter. The filtrate (100 μL) was inoculated into semi-SPF embrocated eggs (10 days old) by adopting the method described by WOAH [40]. The eggs were incubated for 48 h at 37°C and then chilled at 4°C for 24 h. After 24 h of incubation, the blunt end of the eggs was removed using sterilized scissors, and allantoic fluid (AF) was harvested and separated by centrifugation at 10,000 × g for 5 min.
2.5.2. Virus Titration by Heamagglutination (HA) Reaction
The presence of heamagglutinin agents in the AF was confirmed by HA reaction [40]. Saline (100 μL) was dispensed into all wells of U-shaped microtiter plates. AF (100 μL) was added to the 1st column of plates and two-fold dilution was performed up to the 11th column. The 12th column was used as the negative control. RBCs (0.7%, 100 μL) were then added to each well and incubated at 37°C for 30 min. The tubes were observed for agglutination reaction and button formation. HA titer was expressed in terms of log2.
2.6. Serological Detection of NDV
2.6.1. Detection of NDV Exposure by ELISA
The collected serum samples were further investigated by ELISA (IDEXX kit) for the detection of NDV infection.
2.6.1.1. ELISA Procedure
The serum sample was diluted to 1:500 by transferring 1 μL of the serum sample to 449 μL of dilution buffer. An NDV-coated plate was obtained, and the seal was removed. The negative and positive controls were dispensed (100 μL) in duplicates. Prediluted serum sample (100 μL) were added to each well (except the negative and positive controls) and incubated for 30 min at 18°C–26°C (± 2 min). The solution was aspirated from each well, and the plate was washed with distilled water (350 μL) five times. Before drying the plate, 100 μL of conjugate was added to each well and incubated for 30 min at 18°C–26°C (± 2 min). This was followed by the removal of liquid from the wells of the plate using an automatic plate washer and washing the plate with distilled water (350 μL) five times. Substrate solution (100 μL) was added to each well and incubated for 15 (± 1) min at 18°C–26°C. Finally, 100 μL of stop solution was added and absorbance was recorded at 650 nm.
Interpretation is as follows: negative S/P ≤ 0.20; positive S/P > 0.20.
The infection of NDV was detected by comparing the limit of vaccine titer in different weeks with the observed titer during the investigation (Table 2).
| S/no | Limit of vaccine titer at different ages | ||||
|---|---|---|---|---|---|
| DOC | 5–6 weeks | 10–12 weeks | 22–55 weeks | Older than 55 weeks | |
| Breeder | 1500–4000 | 3500–6000 | 5000–8000 | 7000–9000 | 6000–8000 |
| Layer | 3000–4000 | 3000–4000 | 4000–6000 | 7000–9000 | 6000–8000 |
| Broiler | 4000–7000 | 500–2000 | |||
- Abbreviation: DOC, day-old chicks.
2.7. Biological Characterization
2.7.1. Intracerebral Pathogenicity Index (ICPI) Assessment
AF harvested from inoculated eggs showing HA titer > 1/16 (HA titer > 24) was diluted 1/10 in PBS, and 0.05 mL was injected (intracerebral space) in 10 chicks (day-old). The inoculated chicks were observed with an interval of 1 day for 8 days, and then, the score of the birds was recorded according to OIE 2012. At each observation, the birds are scored as follows: 0 if normal, 1 if sick, and 2 if dead. An ICPI score ≥ 0.70 was considered pathogenic [41].
2.7.2. Mean Death Time (MDT) of the Minimum Lethal Dose (MLD)
The MDT was investigated for 10-day-old embrocated eggs. Serial dilutions of allantois fluid of each local virus isolate were prepared, and then, 0.1 mL of each dilution (10–6, 10–7, 10–8, and 10–9) was inoculated into the allantois cavity of eggs. The highest dilution at which all inoculated embryos died soon after was noted, and the MLD and MDT were calculated [42, 43].
2.8. Molecular Detection
2.8.1. RNA Extraction
NDV RNA was extracted from the harvested allantois fluid after 48 h of incubation at 37°C. The fluid was centrifuged at 8000 × g for 5 min, and NDV RNA was extracted according to the manufacturer’s instructions (Life Technologies). Viral RNA was extracted from HA-positive samples using a TRIzol RNA extraction kit (Catalog Numbers 15596026 and 15596018). After thawing, 250 μL of HA-positive allantois fluid was mixed with 750 μL of TRIzol reagent (1:3 ratio) and incubated at room temperature for 10 min. After incubation, the mixture was centrifuged at 12,000 g for 10 min. The supernatant was transferred to a fresh 1.5 mL tube and 250 μL chloroform was added and incubated at room temperature for 10 min. The mixture was centrifuged at x12,000 g for 15 min at 4°C. The aqueous phase containing RNA was transferred into a fresh tube, and then, chilled isopropanol (350 μL) was added into the tube and incubated at room temperature for 10 min to precipitate RNA. Centrifugation was performed for 10 min at 10,000 g at 4°C. The supernatant was discarded, and 750 μL of chilled ethanol was added to the tube. Final centrifugation was performed for 5 min at 7500 g to wash the RNA pellet. This step was repeated three times. The RNA pellet was dissolved in RNase-free water and stored at −80°C. The extracted RNA was quantified using a NanoDrop spectrophotometer. The final extract was dissolved in RNase-free water (Promega) and used for RT-PCR.
2.8.2. RT-PCR
2.8.2.1. RT-PCR Reaction Mixture
Molecular identification was carried out by RT-PCR targeting the Fusion gene of NDV. The RT-PCR reaction was performed in 50 μL reaction mixture containing reagents 10 μL AMV/Tft 5x reaction buffer, 0.2 mM/μL of dNTPs, 1 mM MgSO4, 0.1 μM of forward primer (4306F) 5′GACCGCTGACCACGAGGTTA′3, reverse primer (5005R) 5′AGTCGGAGGATGTTGGCAGC′3, 0.1 U/μL of AMV reverse transcriptase, Tft DNA polymerase, and 8.42 ng/μL RNA template. After the addition of all reagents, the volume was raised up to 50 μL with RNase-free water.
2.8.2.2. RT-PCR Conditions
RT-PCR was carried out to synthesize cDNA at 45°C for 60 min followed by PCR. The 30 cycles of PCR consisted of an initial denaturation at 95°C for 1 min, cycling denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 3 min with the final extension of 72°C for 10 min [44]. The PCR products (700 bp) were visualized on a 2% agarose gel after staining with ethidium bromide.
2.9. Molecular Characterization
2.9.1. Nucleotide Sequencing
- 1.
IsolateChicken/Pakistan/691, NDV accession NoMN096605.
- 2.
IsolateChicken/Pakistan/633, NDVaccession NoMN096604.
- 3.
IsolateChicken/Pakistan/697, NDV accession NoMN096603.
- 4.
IsolateChicken/Pakistan/688, NDV accession NoMN096606.
2.9.2. Phylogenetic Analysis
The sequence of previously characterized reference strains of Asia, the Middle East, and isolates consisting of Genotypes (I–IX) from other parts of the world were retrieved from GenBank (http://www.ncbi.nlm.nih.gov). The sequence data was imported into Clustal W for alignment. The aligned file was imported into Gene DOC for multiple sequence alignment. The multiple sequences alignment file was created and imported into MEGA 11 software for phylogenetic analysis. The phylogenetic analysis was done by the maximum likelihood method [45, 46]. The substitution mutation analysis of the Fusion gene cleavage site was done by comparing the sequence of local isolates and the sequence of reference strains.
2.9.3. Determination of Amino Acid Sequence at Cleavage Site of Fusion Protein
For molecular pathotyping of NDV, the nucleotide sequence in the Fusion gene cleavage site was translated into amino acids using MEGA Version 11, and the amino acid sequence at the Fusion protein cleavage site was determined and compared with the amino acid sequence of previously published reference isolates retrieved from the GenBank. Pathotyping was done by identifying the molecular pathogenic factors (R, R Q, R, R, and ↓F) present in the targeted cleavage site of Fusion protein at the position of 112–117.
3. Results
3.1. Clinical Findings
Mortality rates of up to 40% were observed in vaccinated birds, and even these birds had adequate vaccine titers, as indicated in Table 3. The affected birds were found to have clinical signs in the respiratory and nervous system. The clinical signs included difficulty in breathing, coughing, sneezing, nasal discharge, swollen eyes, and greenish diarrhea. The birds succumbed to death a week after clinical manifestation, with the following postmortem lesions.
| Flocks | Age (weeks) | Examined bird | Birds status | No. of birds show lesions | Previous vaccinea | Mortality | Observed clinical signs/lesions |
|---|---|---|---|---|---|---|---|
| Titer | |||||||
| Broiler | 3–7 | 141 | Morbid/dead | 72 | 4–6 log2 | 25%–40% | 1 respiratory |
| Flock 1 | 5 | 21 | Morbid/dead | 13 | 5.3 log2 | 31 | • Difficulties in breathing |
| Flock 2 | 4.1 | 13 | Morbid/dead | 7 | 4.9 log2 | 37 | • Nasal discharge |
| Flock 3 | 6 | 23 | Morbid/dead | 9 | 5.7 log2 | 28 | • Swollen eyes |
| Flock 4 | 5.2 | 18 | Morbid/dead | 11 | 5.3 log2 | 26 | • Swelling of the head |
| Flock 5 | 5 | 22 | Morbid/dead | 8 | 5.7 log2 | 25 | • Difficulties in breathing |
| Flock 6 | 4 | 24 | Morbid/dead | 11 | 4.1 log2 | 40 | • Sleepy appearance |
| Flock 7 | 4.6 | 20 | Morbid/dead | 13 | 5.1 log2 | 27 | 2 production |
| • Poor weight gain | |||||||
| 3 neurological disorder | |||||||
| Tremors | |||||||
| • Paralysis of wings and legs | |||||||
| Muscle | |||||||
| • Dizzy | |||||||
| 4 hemorrhagic | |||||||
| • Trachitis | |||||||
| • Intestine | |||||||
| • Proventriculus | |||||||
| • Ceacal tonsil | |||||||
| Layer | 8–110 | 122 | Dead | 61 | 7–8.5 log2 | 10%–16% | 1 respiratory |
| Flock 1 | 110 | 25 | Morbid/dead | 10 | 7.6 log2 | 13 | • Difficulties in breathing |
| Flock 2 | 25 | 18 | Morbid/dead | 11 | 8.1 log2 | 10 | • Nasal discharge |
| Flock 3 | 14 | 31 | Morbid/dead | 17 | 8.5 log2 | 16 | • Swollen eyes |
| Flock 4 | 55 | 28 | Morbid/dead | 14 | 7.3 log2 | 13 | • Swelling of the head |
| Flock 5 | 98 | 20 | Morbid/dead | 9 | 7 log2 | 10 | • Difficulties in breathing |
| • Sleepy appearance | |||||||
| 2 production | |||||||
| • Low egg production | |||||||
| 3 neurological disorder | |||||||
| Tremors | |||||||
| • Paralysis of wings and legs | |||||||
| Muscle | |||||||
| • Dizzy | |||||||
| 4 hemorrhagic | |||||||
| • Trachitis | |||||||
| • Intestine | |||||||
| • Proventriculus | |||||||
| • Ceacal tonsil | |||||||
| Breeder | 10–80 | 107 | Dead | 38 | 7–8.3 log2 | 8%–12% | 1 respiratory |
| Flock 1 | 34 | 20 | Morbid/dead | 7 | 8.3 log2 | 10 | • Difficulties in breathing |
| Flock 2 | 10 | 28 | Morbid/dead | 11 | 8 log2 | 8 | • Nasal discharge |
| Flock 3 | 80 | 34 | Morbid/dead | 15 | 7.8 log2 | 12 | • Swollen eyes |
| Flock 4 | 53 | 25 | Morbid/dead | 5 | 7.3 log2 | 8 | • Swelling of the head |
| • Difficulties in breathing | |||||||
| • Sleepy appearance | |||||||
| 2 production | |||||||
| • Low egg production | |||||||
| 3 neurological disorder | |||||||
| Tremors | |||||||
| • Paralysis of wings and legs | |||||||
| Muscle | |||||||
| • Dizzy | |||||||
| 4 hemorrhagic | |||||||
| • Trachitis | |||||||
| • Intestine | |||||||
| • Proventriculus | |||||||
| • Ceacal tonsil | |||||||
| Total | 370 | 171 | |||||
- aRetrieved from the history of the investigated flock.
3.2. Postmortem Findings
During necropsy, severe NDV-associated gross lesions such as congestion of the trachea, petechial hemorrhages in the intestine and proventriculus, congested lungs, swollen kidneys, and necrotic foci in the spleen were observed. The observed lesions are indicative of infection with the velogenic type of NDV (Figures 1 and 2).
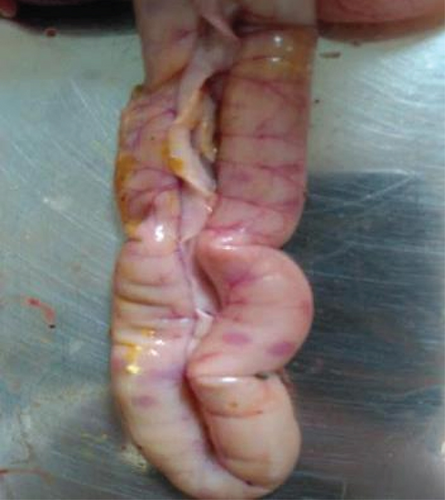

A total of 141 broiler birds, 122-layer birds, and 107 breeder birds were subjected to necropsy. Of these, 72 broilers, 61 layers, and 38 breeder birds exhibited clear postmortem lesions consistent with NDV infection. The mortality rate was 25%–40%, 10%–16%, and 8%–12% in broilers, layers, and breeders, respectively. The vaccine titer (presented by the farmer) in affected birds was found 4–6 log2 in broiler flocks, 7–8.5 log2 in layer, and 7–8.3 log2 in breeder flocks (Table 3). Despite the significant HI titer, infection in vaccinated flocks indicates the circulation of genetic variants in the fields.
3.3. Serological Detection of NDV Infection
3.3.1. ELISA Titer Greater Than the Limit of Vaccine Titer
During the investigation of serum samples, infection with NDV was detected in vaccinated flocks by observing maximum CV values and ELISA titers greater than the limit of the vaccine titer. The higher antibody titer compared to the vaccine titer (Table 4) suggested NDV infection in 368 out of 877 serum samples collected from sick birds (Figure 3). The serum samples of infected birds showed an ELISA titer greater than the limit of the vaccine titer, indicating infection.
| Flock | Tested serum samples | Positive by ELISA | Prevalence % | Mini CV% | Max CV% | Max titer | Indications by | |
|---|---|---|---|---|---|---|---|---|
| CV% | ELISA titer | |||||||
| Breeder | 387 | 105 | 27% | 36.6 | 42.1 | 25020 | Exposure | Exposure |
| Layer | 256 | 116 | 45% | 39.8 | 52.3 | 20732 | Exposure | Exposure |
| Broiler | 234 | 147 | 62% | 33.4 | 37.3 | 4173 | Exposure | Exposure |
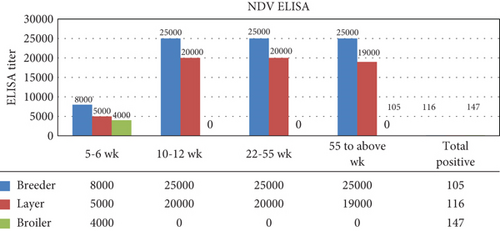
The observed antibody titer in breeder flocks was up to 25,020 with a minimum CV% value of 36.3 to a max CV% value of 42.1. Both the ELISA titer and CV% indicated NDV infection in the folks. In the layer flock, the maximum ELISA titer was found 20,732 with CV% values of minimum 39.8 to max 52.3. The maximum ELISA titer in broiler flocks was 4173 with a CV% value minimum of 33.4 to a maximum of 37.3. Both the ELISA titer and CV% values in layer and broiler flocks also indicate the infection of NDV in tested flocks. The infection of NDV in breeder flocks was found to be 27%, layer 45%, and broiler 62% by ELISA (Table 4).
3.3.2. Detection of Higher ELISA Titer in the Affected Flock
The elevated antibody titer was quantified using ELISA. The ELISA titer at 5–6 weeks in the breeder, layer, and broiler flocks was greater than the vaccine titer provided by the manufacturer (breeder < 8000, layer < 5000, and broiler < 4000). ELISA titers at 10–12 and 22–25 weeks in the breeder and layers were greater than the vaccine titer (breeder < 25,000 and layer < 20,000). The ELISA titer above 55 weeks in the breeder and flock also indicated infection (breeder < 25,000; layer < 1900) (Figure 3). All observed titers in different flocks suggested NDV infection in the investigated flocks.
3.4. Molecular Diagnosis
3.4.1. Molecular Diagnosis of NDV by RT-PCR
The results of RT-PCR showed a higher prevalence of NDV in the selected area. NDV infection was confirmed by RT-PCR in AF collected after 48 h of incubation. Out of 167 HA-positive AF, 66 AF samples were found positive for NDV by amplifying the Fusion gene which produced the expected band size of 700 bp (Figure 4). The RT-PCR product was subsequently used for the molecular characterization of NDV by gene sequencing.
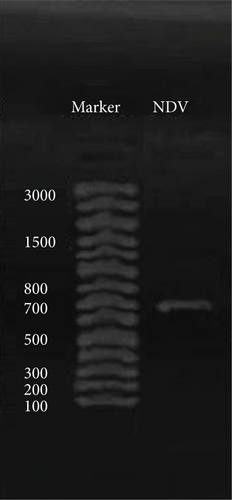
3.4.2. Prevalence of NDV Detected by RT-PCR
NDV infection was detected by Fusion gene-based RT-PCR in the affected flocks. Different prevalence percentages of NDV were detected in previously vaccinated (multiple dose) infected flocks. The prevalence of NDV was 21% in broiler flocks, 17% in layers, and 15% in breeders (Table 5).
| Bird types | Investigated bird status | Vaccine shots up to culling | Vaccine strain | Total sample collected | Positive by culture and Fusion gene PCR | % of positive |
|---|---|---|---|---|---|---|
| Broiler | Morbid/dead | 4 times | Genotype II | 141 | 29 | 21 |
| Layer | Morbid/dead | 9 times | Genotype II | 122 | 21 | 17 |
| Breeder | Morbid/dead | 6–9 times | Genotype II | 107 | 16 | 15 |
| Total | 370 | 66 | 18 | |||
3.4.3. Phylogenetic Analysis of NDV Based on Fusion Gene
Phylogenetic analysis of the partial Fusion gene sequence of the selected isolates revealed high diversity among NDV field isolates and vaccine strains (Genotype II) used in the southern region. All selected local isolates (e.g., Chicken/Pakistan/691 NDV with the accession No. MN096605; Chicken/Pakistan/633 NDV with the accession No. MN096604, and Chicken/Pakistan/688 NDV with Accession No. MN096606) were clustered closely and showed proximity with NDV Genotype VII, which is considered a velogenic NDV type as shown in Figure 5. In contrast, one isolate (Chicken/Pakistan/697/2017 with Accession No. MN096603) was grouped with NDV Genotype II (Figure 5).
3.5. Pathotyping of NDV
3.5.1. Detection of Biological Factors Involve in Pathogenicity
The isolates of NDV (66) obtained from different regions were characterized biologically using methods such as calculation of ICPI in day-old chicks and MDT after inoculation into 10-day-old embryonated eggs [47]. During biological characterization, the ICPI index in day-old chicks was observed from 0.4 to 2 in all isolates, and the MDT was recorded from 48 to > 90 h. During the biological characterization of a total of 66 tested isolates, 57 isolates scored an ICPI index of 1.7–2 with an MDT of 48–53 h and were classified as velogenic. The other nine isolates scored an ICPI index of 0.4–0.5 with the MDT > 90 h and were classified as a lentogenic type. Based on ICPI and MDT, 57 isolates were considered velogenic, and nine isolates were characterized as lentogenic (Table 6).
| Region | No. of isolates | ICPI index | MDT (h) | NDV | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.2 | 0.3 | 0.4 | 0.5 | 1.6 | 1.7 | 1.8 | 1.9 | 2 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 72 | > 90 | Pathotype | ||
| Karachi | 20 | 0 | 0 | 0 | 0 | 0 | 6 | 4 | 2 | 8 | 10 | 2 | 4 | 2 | 2 | 0 | 0 | 0 | 0 | Velogenic |
| Hyderabad | 9 | 0 | 0 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9a | Lentogenica |
| Theta | 23 | 0 | 0 | 0 | 0 | 0 | 5 | 8 | 3 | 7 | 12 | 6 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | Velogenic |
| Dado | 14 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 2 | 6 | 6 | 1 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | Velogenic |
| Total | 66 | 0 | 0 | 6 | 3 | 0 | 12 | 17 | 7 | 21 | 28 | 9 | 12 | 6 | 2 | 0 | 0 | 0 | 9 | |
- aPrevalence of low pathogenic type.
3.5.2. Detection of Molecular Factors Involved in Pathogenicity
The molecular factors (amino acids) involved in the pathogenicity of the NDV Fusion protein were determined. The amino acid sequence of Fusion protein region 112–117 was compared with the previously published amino acid sequence of isolates retrieved from GenBank (Table 1). All field isolates (such as Chicken/Pakistan/691 NDV, Chicken/Pakistan/633 NDV, and Chicken/Pakistan/688 NDV) show similar velogenic cleavage sites with four basic amino acid residues R, R Q, R, R, and ↓F with phenylalanine (F) at position 117. These sequences have been postulated to be potent molecular determinants for severe velogenic effects during infection [48, 49]. Analysis of the amino acid sequence of the Fusion protein cleavage site showed that all isolates of NDV belong to the virulent type of NDV. Only one isolate, Chicken/Pakistan/697, showed two basic amino acids G, R, Q, G, and R↓L in their cleavage site indicating their lentogenic nature (Table 7).
| Strain | Fusion protein sequence 109–120 | Pathotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fusion protein cleavage motif | Based on Fusion protein cleavage site | ||||||||||||
| Accession no. | 109 | 110 | 111 | 112 | 113 | 114 | 115 | 116 | 117 | 118 | 119 | 120 | |
| EU293914.1 | S | G | G | R | R | Q | R | R | ↓F | I | G | A | Virulent |
| AY741404.1 | S | G | G | R | R | Q | R | R | ↓F | I | G | A | Virulent |
| GU187941.1 | S | G | G | R | R | Q | R | R | ↓F | I | G | A | Virulent |
| FJ436302.1 | S | G | G | R | R | Q | R | R | ↓F | I | G | A | Virulent |
| JX518883.1 | S | G | G | R | R | R | R | R | ↓F | V | G | A | Virulent |
| AY845400.2 | S | G | G | G | R | Q | G | R | ↓L | I | G | A | Avirulent |
| AJ629062.1 | S | G | G | G | R | Q | G | R | ↓L | I | G | A | Avirulent |
| X04719.1 | S | G | G | R | R | Q | K | R | ↓L | I | G | A | Virulent |
| AY935499.2 | S | G | G | R | K | Q | G | R | ↓L | I | G | A | Avirulent |
| HM063422.1 | S | G | G | E | R | Q | G | R | ↓L | I | G | A | Avirulent |
| EF201805.1 | S | G | G | R | R | Q | R | R | ↓F | I | G | A | Virulent |
| JX173098.1 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| KJ450980.1 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| KY693681.1 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| KU365652.1 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| aMN096604 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| aMN096606 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| aMN096605 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| aMN096603 | S | G | G | G | R | Q | G | R | ↓L | I | G | A | Avirulent |
| GQ288388.2 | S | R | G | R | R | Q | K | R | ↓F | V | G | A | Virulent |
| FJ766526.1 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
| AY734534.1 | S | G | G | R | R | Q | K | R | ↓F | I | G | A | Virulent |
- aLocal Pakistani isolates.
3.6. Amino Acid Substitution in Fusion Protein
The substitution mutation in the amino acid sequence of the Fusion protein was determined by reverse transcription of the Fusion gene sequence of a selective NDV isolate. A unique substitution in the amino acid sequence of the Fusion protein at Positions 105 and 107 in Pakistani isolates Chicken/Pakistan/633 NDV (MN096604) and Chicken/Pakistan/691 NDV (MN096605) was observed. Serine (S) was replaced with proline (P) and serine/thymine (S/T) was replaced with alanine (A), while all other reference isolates showed S at position 105 and S/T at position 107 of the Fusion proteins. Another selective isolate Chicken/Pakistan/688 NDV (MN096606) showed a substitution at position 107, where S/T was replaced by A. Only one isolate Chicken/Pakistan/697 NDV (MN096603) showed a similar amino acid pattern with other reference avirulent NDVs. The dotted lines in the box represent identical sequences of amino acid mentioned in the first row; the single alphabet in the box represents differences in amino acid sequences (Table 8).
| The variable amino acid sequence of Fusion protein of NDV field isolate and reference strains | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 20 | 105 | 107 |
| JX173098.1 | M | G | S | K | P | S | R | I | P | E | S | S | M | S | S |
| KJ450980.1 | — | — | — | — | — | — | — | — | A | P | L | — | — | — | |
| KY693681.1 | — | — | P | — | — | — | — | T | — | A | P | L | — | — | — |
| KU365652.1 | — | — | — | — | — | — | — | — | — | A | P | L | — | — | — |
| aMN096605 | — | — | — | — | — | — | — | — | — | A | P | L | — | sP | sA |
| aMN096604 | — | — | — | — | — | — | — | — | — | A | P | L | — | sP | sA |
| aMN096606 | — | — | — | — | — | — | — | — | — | A | P | L | — | — | sA |
| FJ766526.1 | — | D | — | — | — | H | — | — | — | A | P | L | T | — | — |
| GQ288388.2 | — | — | P | — | — | — | — | T | — | A | P | L | T | — | T |
| AY734534.1 | — | — | — | — | S | — | — | — | — | T | P | L | L | — | T |
| EU293914.1 | — | — | — | R | S | — | — | — | T | V | P | L | A | — | T |
| AY741404.1 | — | — | — | R | S | — | — | — | — | V | P | P | V | — | T |
| GU187941.1 | — | — | — | R | S | — | — | T | — | A | H | L | A | — | T |
| FJ436302.1 | — | — | P | S | — | N | V | — | A | P | L | A | — | T | |
| JX518883.1 | — | — | — | Q | S | — | W | — | — | I | F | P | V | V | T |
| EF201805.1 | — | — | P | R | S | — | — | — | V | P | L | T | T | T | |
| AY845400.2 | — | — | — | R | — | — | K | N | — | A | P | M | A | A | T |
| AJ629062.1 | — | — | — | R | — | — | K | N | — | A | P | M | A | A | T |
| aMN096603 | — | — | — | R | — | — | K | N | — | A | P | M | A | A | T |
| X04719.1 | — | — | P | R | — | — | K | N | — | V | P | M | A | A | T |
| AY935499.2 | — | — | — | — | S | — | — | F | — | V | P | L | — | T | T |
| HM063422.1 | — | — | — | R | S | — | — | — | — | V | P | L | A | A | T |
- aLocal isolates
- sAmino acid substitution.
3.7. Distribution of NDV Genotypes in the South Region
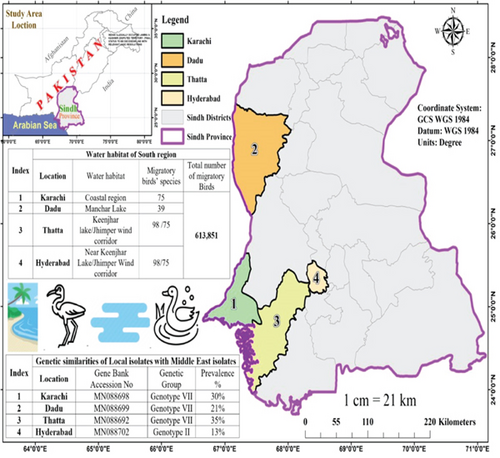
The NDV Genotype VII was found to be prevalent in the targeted district of the southern region (Sindh) with the exception of Hyderabad, where NDV Genotype II was predominant. The prevalence of NDV Genotype VII was detected in all the selected regions including Karachi (30%), Dadu (21%), and Thatta (35%) while Genotype II was prevalent in Hyderabad (13%). The highest prevalence of NDV Genotype VII was recorded in Thatta District, followed by Karachi and Dadu District (Figure 6).
4. Discussion
NDV is endemic in Pakistan and poses a significant threat to the poultry industry [50]. The high mortality in the vaccinated poultry flocks indicates the circulation of the virulent strains in the field. As a mitigation strategy, it is essential to characterize the prevalent strains and to identify the genetic cluster of the NDV in the region. Based on its Fusion gene sequence, various diverse genetic clusters of NDV have been identified [22, 51], which have been further divided into subtypes. The prevalence of diversified genetic groups of NDV V, VI, VII, and VIII has been reported from different geographic regions including Pakistan [37, 52], China, Malaysia, Kazakhstan, Kyrgyzstan [53, 54], and Africa [55]. Among these, Genotypes V, VI, VII, and VIII are considered as most prevalent genotypes in the world while genotype VII has been found a prevalent subtype in Asia [56]. Circulation of VII and other genetic groups have also been reported from different regions of Pakistan [36–38], but molecular epidemiology of the NDV has rarely been reported from the southern region.
Interestingly, the vaccine against Genotype II is commonly used in the southern region of Pakistan with the repeated shots twice per month. Despite extensive vaccination, NDV outbreak in the vaccinated commercial poultry flocks was noted in this region. Although protective HI titers ranging from 4 to 8 log2 were present, a very high mortality rate was observed in the vaccinated flock. The susceptible birds initially showed clinical signs and symptoms related to NDV and then succumbed to death a week later; the presence of severe postmortem lesions in various organs indicated the infection by a virulent strain. The results of the ELISA test confirmed the infection by NDV. This situation indicated a distinct genetic group of the infective agent than the vaccine strain. Molecular characterization and phylogenetic analysis of the selected isolates revealed the presence of a single Genotype VII of NDV in vaccinated flocks. Indeed, all the selected isolates were clustered with Genotype VII. In contrast, the involvement of two genotypes, VI and VII, was reported in 2010 [52], while the lentogenic Type II and velogenic Type VII were reported from different regions of Punjab [37, 57]. Following these reports, another study revealed the prevalence of a single Type VII from different regions of Panjab highlighting an outbreak of this velogenic type [37]. This finding also indicated the replacement of previously reported genotypes (II, IV, and XIII) from 2010 to 2015. This finding is also supported by previous studies which revealed the replacement of Genotype XIII with a new emerging genotype VII [58, 59]. Consequently, Genotype VII was found continually associated with fatal outbreaks in the vaccinated flocks as an endemic type from northern to southern regions. Although its origin is not certainly known, the growing number of reports suggests its transmission from densely populated regions of Punjab.
Punjab is a densely populated region where different types of poultry flocks prevail including GP, breeder, commercial layer, and broiler. On the other hand, the wild birds are crucial in NDV epizootic [58]. Various species of wild birds play a significant role in the transmission of NDV. Some incidence was reported from seven districts of the southern region where wild peacocks have been affected by the virulent type of NDV [36]. Hence, it is assumed that wild birds may be a potential source of virulent NDV transmission in the southern region, because the southern region is a major hub for wild birds where various species of migratory birds are inhabitated during the summer and winter seasons. Particularly the lakes, Kanjher and Manchar, reportedly receive 98 and 39 migratory water bird species [60, 61]. Likewise, the other hotspots in the southern region including Hub Dam, Jhimpir Wind Corridor, and the Coastal wetland received 134, 79, and 75 migratory birds, respectively [62, 63]. As these lakes, dams, and wetlands are of socioeconomic importance where villagers depend on their maximum livelihood; therefore, these may act as a source for the dissemination of disease from wild birds to the backyard followed by commercial poultry. It is of particular interest as earlier reports indicate the presence of Genotype VII in backyard poultry [37]. NDV of Genotype VII has also been associated with the outbreak in the Middle East [64] that may have been reached to the southern region of Pakistan through wild birds as this genotype has also been reported in the wild birds [65]. Hence, a comprehensive investigation is required to investigate the role of water birds in the dissemination of NDV.
The virulent infection in the vaccinated flocks [37, 38] indicated the continuous evolution in Genotype VII and consistent circulation in the fields affecting the flocks vaccinated with Genotypes II and III. All types of flocks in open and controlled houses were found to be affected by the NDV Genotype VII indicating its persistence. The variation between the vaccine strains and the field type poses a great challenge [66, 67]. The emergence and circulation of new genetic groups among vaccinated flocks have been reported earlier [57, 68]. The emergence of diverse genotypes and their significant threats was also recognized by Miller et al. [69]. The spread of Genotype VII and its significant impact on vaccination programs was noted globally, especially in South America [70] and the Far East [71, 72]. Recently, vaccine failure leading to a change in the vaccination program has also been recommended in Egypt [73]. These findings raised the questions against conventional vaccines and suggested changes in vaccination programs worldwide.
For a long time, it was commonly accepted that the conventional NDV vaccines (G II) are effective in controlling NDV infection; however, experimental studies reported a failure in preventing infection caused by a completely different genotype circulating in the field [74–77]. Recent studies claim that vaccines based on genetically related types provide better protection against the GVII field virus compared with Genotype II NDV vaccines [73, 74, 77–80]. The phylogenic gaps between the vaccine strain and field-challenged virus were observed during the investigation which may be attributed to a lack of protection in the vaccinated flocks.
The failure of protection for the birds vaccinated with the live vaccine demands a nationwide comprehensive molecular epidemiological investigation on a regular basis. This will aid in making the vaccination program effective.
This study also revealed the pathogenic potential of the circulating NDV which was attributed to the amino acid sequence of the cleavage motif in Fusion protein [31, 33, 81, 82]. The amino acid analysis of the Fusion protein cleavage site of the selected NDV isolates had polybasic amino acids from positions 112–117 (R112R113Q114K115R116 ↓F117) with F at Position 117. The presence of these factors in the Fusion proteins indicates that all isolates belonged to the highly pathogenic velogenic type. The sequence analysis of the Fusion protein confirmed that the outbreak of this velogenic type had a pathogenic identity with previously reported velogenic types isolated from 2012 to 2013 in Panjab and other regions of Pakistan [38, 57]. The in vivo pathogenicity assays, ICPI and MDT, also classified the isolate into two pathotypes, velogenic and lentotype.
Fusion protein plays a pivotal role in NDV virulence as some of its substituted amino acid mutations have an association with the pathogenic potentials by increasing the fusogenic ability of the NDV [83]. Some point mutations in Fusion protein have been reported in the isolates from Pakistan [37]; however, their role in pathogenicity has not been fully elucidated. A unique substitution mutation in the amino acid sequence of Fusion protein was found at Positions 105 and 107 where S was substituted by P in isolates of Chicken/Pakistan/633/2016 (NDV), Chicken/Pakistan/691/2016 (NDV), and another isolate Chicken/Pakistan/688/2017 (NDV), where S/T amino acid was substituted by A at the position of 107. These mutations indicate the genetic diversity of selected isolates. The unique substitution mutation of amino acid in the Fusion protein of selected isolates may be another virulent factor responsible for increasing the pathogenic potentials and vaccine failure with fatal consequences in vaccinated flocks. In previous studies, it was noted that a mutation in the Fusion protein cleavage site potentially affects the pathogenicity of NDV [84]. Such findings need detailed investigations.
The epizootic and favorable situation for NDV in Pakistan adds a potential risk for sweeping of current genotype with a newly emerging genotype which exacerbates the vaccination program and risk of repeated outbreaks. The replacement of previously prevalent Genotype II with Genotype VII NDV throughout Pakistan raises the question of improving the preventive measures in commercial poultry.
Addressing these issues requires a comprehensive approach including proper vaccine selection based on the current molecular epidemiological situation, storage and maintenance of cold chain, handling of vaccine, and administration of vaccine and measures to improve overall flock health. Additionally, ongoing surveillance of circulating NDV strains and vaccination efficacy can help inform decision-making and control strategies.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This project was funded by the University of Karachi.
Open Research
Data Availability Statement
The data associated with this manuscript can be provided on reasonable request.




