Whole-Genome Sequencing and Functional Analysis of Klebsiella Strain mr-1S Originating From Macrobrachium rosenbergii
Abstract
Background:Klebsiella mr-1S, a newly emerging pathogen that poses a substantial threat to public health by inducing severe infectious diseases, has been detected in Macrobrachium rosenbergii for the first time in our prior investigation. Given its potential for environmental adaptation and genomic evolution, this study aims to delve into these aspects to better understand its pathogenicity and evolutionary trajectory.
Methods: In this study, the Klebsiella isolates were subjected to routine cultivation in a tryptic soya broth (TSB) medium. Subsequently, the genomic DNA of each isolate was individually extracted and analyzed. Prior to genomic sequencing, the integrity and concentration of the DNA samples were meticulously evaluated to ensure the accuracy and reliability of the subsequent sequencing process.
Results: The genomic sequence of Klebsiella mr-1S was deciphered, revealing a length of 5,143,806 base pairs with a GC content of 54.97%. Remarkably, the genome encompasses a multitude of putative mobile genetic elements (MGEs), including 43 genomic islands (GIs) and 2 prophages. These elements confer upon the bacterium crucial adaptive attributes such as resistance, virulence, and metabolic capabilities. Notably, the identification of prophage-associated clusters originating from the genus Pseudomonas suggests a potential horizontal gene transfer (HGT) mechanism mediated by phages within Pseudomonas, highlighting the complex genetic interactions between different bacterial species.
Conclusion: In addition, the presence of two genes encoding CRISPR-Cas proteins within the Klebsiella mr-1S genome indicates the existence of a functional CRISPR-Cas system in this bacterium. This finding implies that during its evolutionary history, Klebsiella mr-1S may have developed mechanisms to evade host immune recognition, thereby facilitating efficient HGT and enhancing its adaptability and survival capabilities in diverse environments. Overall, this study provides novel insights into the environmental adaptability and genomic plasticity of Klebsiella from Macrobrachium rosenbergii, underscoring its potential as an emerging pathogen in aquatic ecosystems and paving the way for future research on its pathogenic mechanisms and potential control strategies.
1. Introduction
Klebsiella spp., a member of the family Enterobacteriaceae, are nonmotile, Gram-negative bacilli with a capsule. Klebsiella pneumoniae, a common opportunistic Gram-negative pathogen, is widely found in human and animal intestines and the natural environment [1, 2]. Its cell wall has a thin peptidoglycan layer, a thick outer membrane and capsule; most strains have fimbriae but no spores or flagella [3]. In livestock farming, researchers have isolated this bacterium from poultry (chickens, ducks), livestock (pigs, dairy cows, goats, sheep), aquatic animals (carps), and their farming environments [4]. Nevertheless, this opportunistic pathogen can precipitate a spectrum of infectious diseases, including pneumonia [5], urinary tract infections [6], septicemia [7], and wound infections [8]. In recent years, the escalating abuse of antimicrobial agents has propelled the rise of antimicrobial resistance in Klebsiella species, posing a grave threat to global public health and complicating clinical therapeutic decision-making [9]. According to the 2023 report from China’s Antimicrobial Resistance Surveillance Network, Klebsiella has emerged as the second most prevalent clinical isolate, trailing only Escherichia coli, with its drug resistance and incidence rates exhibiting a steady upward trajectory [10]. Klebsiella spp. possess diverse virulence factors, including siderophores, lipopolysaccharides, phenol oxidase, and multiple AMR genes, which greatly reduce the available therapeutic options after infection [11, 12]. Klebsiella spp. significantly curtails the available therapeutic options for infections. Consequently, a comprehensive understanding of the biological characteristics and genomic features of Klebsiella spp. is of paramount importance for optimizing its role as a clinical therapeutic agent.
Klebsiella spp. bacteria are characterized by drug resistance, limiting treatment options for diseases they cause. The spread of transmissible genetic nucleotides, acquisition of resistance genes via horizontal gene transfer (HGT), and dissemination of virulence factors drive bacterial drug resistance; the evolution of virulence and resistance traits is essential for pathogen survival and reproduction [13, 14]. The virulence of Klebsiella spp. is modulated by multiple virulence factors, including the iron uptake-related genes FimH, rmpA, uge, and kfu. There are few reports on Klebsiella spp. isolation. However, a recent study by Sun et al. [15] identified the adaptability of this novel K. pneumoniae strain and its new roles in different ecological niches.
Next-generation sequencing (NGS) is increasingly recognized as a potent instrument in microbiology, enhancing the identification of genes that confer resistance to antimicrobial agents and encode virulence determinants, as well as those linked to their underlying genetic processes [16]. Notably, whole-genome sequencing (WGS) has been extensively applied across the antimicrobial resistance domain, encompassing the innovation of new antimicrobials, the establishment of surveillance networks in both human and veterinary medicine, the examination of resistance evolution in real-time across diverse scenarios, and the advancement of diagnostic assays [17]. Employing molecular typing and analysis of microbial genomic diversity through the NCBI database, it was confirmed that the bacterium in question was Klebsiella, with the 16S rRNA gene sequence (1539 nucleotides in length) exhibiting a close match to the entries within the NCBI database (https://www.ncbi.nlm.nih.gov/). A phylogenetic tree was delineated using the maximum likelihood approach, with the 16S rRNA gene segment of other Klebsiella strains serving as a comparative benchmark (Figure 1).
The primary aims of this research were threefold: (1) to delineate the genomic characteristics of Klebsiella isolates for sequencing; (2) to elucidate the genomic sequences of these Klebsiella isolates, identifying mobile genetic elements (MGEs) and genes pertinent to virulence and resistance within the Klebsiella genome; and (3) to dissect the phylogenetic relationships among Klebsiella isolates. The study’s findings are poised to address previous deficiencies in the genomic understanding of Klebsiella strains of canine origin and to augment our comprehension of the evolutionary and pathogenic mechanisms of emerging pathogens on a global scale.
2. Materials and Methods
2.1. Isolation and Culture Conditions of Klebsiella
Klebsiella strains were procured from multiple hatcheries located in Huzhou City, within the Zhejiang Province, during the spring breeding season of the Macrobrachium rosenbergii in the years 2021–2023. These strains were subsequently cryopreserved at a temperature of −80°C within the National Genetic Breeding Center for M. rosenbergii, Zhejiang Institute of Freshwater Fisheries. The Klebsiella isolates underwent routine cultivation in tryptic soya broth (TSB) medium, which was adjusted to a pH of 7.2 and supplemented with 0.5% sodium chloride (NaCl), sourced from Pernosai Wuhan, China. The cultivation process was conducted at a controlled temperature of 37°C under aerobic conditions, with the culture flasks subjected to constant agitation at a rate of 175 revolutions per minute.
2.2. Sequencing Assembly and Annotation of Genomes
The genomic DNA of Klebsiella isolates was individually extracted using the TIANamp Bacterial DNA kit, a product of Beijing Tiangen Biochemical Technology Co., Ltd., following the protocol provided by the manufacturer. The integrity and concentration of the DNA samples destined for genomic sequencing were meticulously assessed, adhering to the methodologies detailed in the scholarly literature [18]. The Klebsiella genomes were subjected to sequencing on the Illumina HiSeq X Ten platform, employing three distinct DNA samples for each Klebsiella isolate. For the assembly of sequences, gene prediction, and Clusters of Orthologous Groups (COG) of proteins, the software utilized mirrored the configurations delineated in our cited references [19], employing the parameters as originally set.
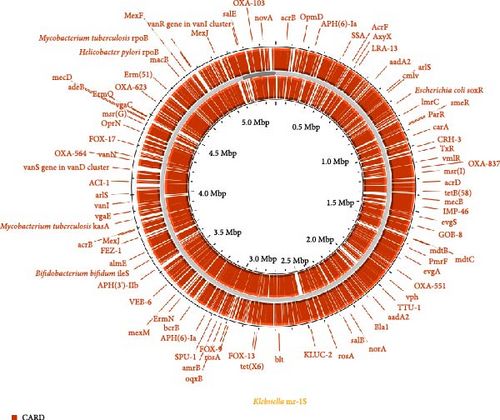
For the identification of virulence-associated genes and resistance-associated genes, the Virulence Factor Database (VFDB), accessible at http://www.mgc.ac.cn/VFs, and the ARGs database, which can be found at http://arpcard.mcmaster.ca, was leveraged. These databases were integrated with the CARD database, accessible at http://arpcard.mcmaster.ca, for the detection of the aforementioned genes [20].
2.3. Comparative Genomic Analysis
The Klebsiella genome was analyzed for MGEs, encompassing genomic islands (GIs), prophages, integrons (Ints), and insertion sequences (ISs), as well as CRISPR-Cas systems, utilizing the identical bioinformatics software and default settings as delineated in our recent publications. Furthermore, the identification of conserved genes shared across Klebsiella isolates and the delineation of strain-specific genes within individual genomes were accomplished using the same software suite and default parameters [21].
A phylogenetic dendrogram was established utilizing the 10 most analogous Klebsiella strains. For the creation of the phylogenetic, genomic dendrogram, single-copy orthologs common to all genomes were deduced employing the OrthoFinder software suite (version 2.5.5). The sequences of each individual single-copy orthologue were then juxtaposed independently using the mafft algorithmic tool (version 7.520). Maximum likelihood dendrograms (with 1000 bootstrap replicates) were generated through a Perl script available at https://github.com/nylander/catfasta2phyml and concurrently with the IQ-Tree software (version 2.2.3). In an analogous fashion, a phylogenetic dendrogram was also derived from the recognized prophage sequences, utilizing the MEGA software platform (https://www.megasoftware.net). The seven conserved core genes characteristic of Klebsiella isolates (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) underwent multilocus sequence typing (MLST) analysis and were subsequently compared against the MLST repository (https://cge.food.dtu.dk/services/MLST/) [22]. The analyses were executed accordingly.
3. Result
3.1. Genome Annotation of the Klebsiella mr-1S Strain
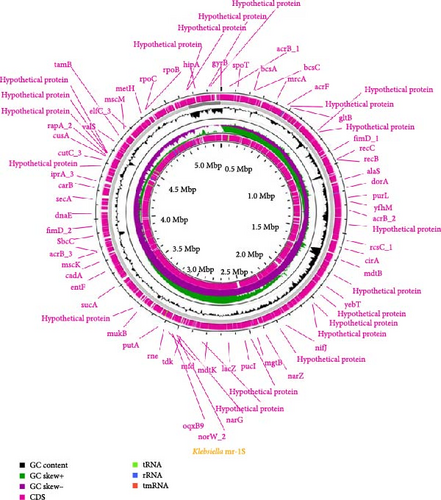
The genomic sequence of the Klebsiella mr-1S strain, which was derived from M. rosenbergii was elucidated utilizing the Illumina sequencing platform. The extant genome encompasses 5,398,092 nucleotides with a GC content of 54.97%. The annotation of the genome has identified a total of 5182 genes, which correspond to 5079 coding sequences (CDSs) and 103 RNA molecules. This includes 23 genes encoding for 16S, 23S, and 5S rRNAs, as well as 80 distinct predicted tRNAs (Figure 1). The genomic analysis has characterized a significant number of subsystems related to protein and carbohydrate metabolism (Figure 2). These encompass genes that participate in processes such as carbohydrate metabolism, aminoglycan metabolism, the genetic transfer of protein information, and the metabolism of cofactors, vitamins, and lipids. Additionally, the genome contains numerous genes associated with glycan metabolism, immune regulation, and transmembrane transport mechanisms.
3.2. Functional Annotation
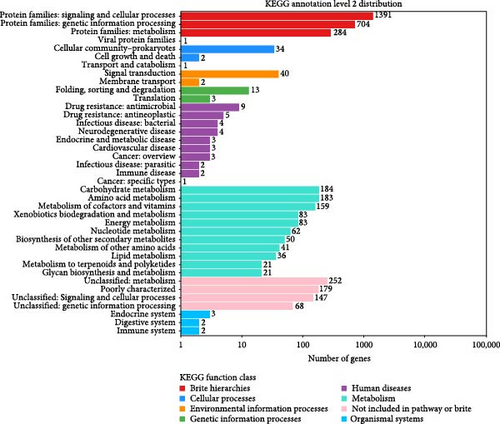
A total of 4804 protein-coding genes, which constitute 92.71% of the total identified protein-coding genes, have been assigned putative functions based on their immediate homology within the COG classification. The genes related to carbohydrate transport and metabolism (comprising 493 open reading frames, ORFs), translation (with 228 ORFs), and transcription (encompassing 215 ORFs) are recognized as the most prevalent functional categories within the COG and KEGG classification. The distribution of these genes across the various COG and KEGG functional categories is detailed and summarized in Figures 2 and 3.

3.3. Phylogeny and Classification
Based on the 16S rDNA sequence analysis, the phylogenetic tree delineated that the Klebsiella mr-1S strain exhibited a higher degree of similarity to the Klebsiella quasipneumoniae subsp. similipneumoniae strain 07A044, as compared to other strains within the Klebsiella genus. Furthermore, genome-to-genome comparative analyses between the Klebsiella mr-1S strain and its closely related strains were conducted utilizing the genome-to-genome distance calculator (GGDC). The DNA–DNA hybridization (DDH) metric was identified as the most reliable indicator for distinguishing between species. Logistic regression analysis, applied to three distinct formulas, predicted a high probability of identifying a DDH value exceeding 70%. Specifically, it was determined that the Klebsiella mr-1S strain had a DDH value of greater than 96% when compared with the Klebsiella quasipneumoniae subsp. similipneumoniae strain 07A044. Subsequent analyses, integrated with phylogenetic methodologies based on the 16S rDNA sequences, substantiated the classification of the Klebsiella mr-1S strain as a member of the Klebsiella (Figure 4).

3.4. Comparative Analysis of Virulence Genes
The Klebsiella mr-1S strain was scrutinized for the presence of virulence-related genes. Examinations were conducted using the VFDB, an extensive resource available at http://www.mgc.ac.cn/VFs/. The VFDB is maintained by the NHC Key Laboratory of Systems Biology of Pathogens (National Institute of Pathogen Biology, Beijing, China).
Upon analysis, it was observed that nearly 30 genes typically associated with virulence in enterococci were absent from the Klebsiella mr-1S strain. Notably, the esp. gene, which encodes the enterococcal surface protein and is a known virulence factor, was not detected. However, several other genes were identified, including ebpA (DTX73_01685), ebpB (DTX73_01690), ebpC (DTX73_01695), srtC (DTX73_01700), ecbA (DTX73_00685), and efaA (DTX73_03830), which are considered to contribute to the pathogenic potential of the strain.
3.5. Genes for Antibiotic Resistance
The Klebsiella mr-1S strain was found to harbor genes linked to antibiotic resistance and the detoxification of toxic substances. These genes are homologous to those known to confer resistance to aminoglycosides, as well as to those associated with MLS resistance (a group of antibiotics including macrolides, lincosamides, and streptogramin B).
Furthermore, the pathogen genome annotation and analysis pipeline (PGAAP) and rapid annotation using subsystem technology (RAST) annotation systems were instrumental in identifying an additional 52 genes that may be implicated in virulence and defense mechanisms. These systems provide a comprehensive analysis of the genomic content, allowing for the detection of genes that could play a role in the pathogenicity, disease progression, and defensive strategies of the Klebsiella mr-1S strain. This discovery underscores the complexity of the strain’s genetic makeup and its potential to adapt to various environmental challenges, including the presence of antibiotics and other harmful compounds.
3.6. Predicted MGEs Protophage
In the present investigation, a single prophage harboring a quintet of genes was discerned within the genomic sequence of the Klebsiella mr-1S strain. The deduced prophage encompasses genetic constituents pertinent to the processes of integration and excision, capsid and tail assembly, cellular lysis, as well as DNA alteration and immunity mechanisms.
3.7. ISs
The Klebsiella mr-1S strain’s genomic sequence harbors a singular IS, spanning 834 base pairs, which is classified under the IS5 family and encodes a protein from the short-chain dehydrogenase/reductase (SDR) family. This suggests that the IS functions as a transport molecule.
3.8. CRISPR-Cas
The Klebsiella mr-1S strain is endowed with a pair of CRISPR box arrays (n = 2). Furthermore, a variety of repetitive sequences were identified within the CRISPR box array of the Klebsiella mr-1S strain’s genome. These findings offer circumstantial evidence suggesting that while the strain’s adaptive immune system may be inactive, HGT could be operational in the isolates of Klebsiella mr-1S.
4. Discussions
Antibiotics are extensively employed in human healthcare, veterinary medicine for food-producing animals, and agricultural practices, with the dual objectives of preventing and combating infections. In aquaculture, the indiscriminate use of antibiotics is prevalent, and it is often not subject to rigorous regulatory oversight; this practice leads to the dissemination of these substances in aquatic environments and sediments, thereby promoting the emergence and spread of antibiotic resistance within aquatic microbial communities [23]. Consequently, these bacteria may function as reservoirs of resistance genes, posing a significant threat to public health.
In this study, we present the genome sequence of a Klebsiella strain, analyzing its phenotypic characteristics and pathogenicity. Our work highlights the correlation between genome sequences and the biological traits of this species. Through WGS, we achieve a more detailed analysis of its genetic structure. For instance, phylogenetic analysis of core genes offers compelling evidence for reclassifying this novel variety as a potential new species. The whole-genome results not only underscore the challenges and potential inaccuracies of identifying Klebsiella species based on single or limited genes but also emphasize the importance of comprehensive genome analysis for accurate taxonomy [24]. The accuracy of classification is closely related to the breadth and quality of the reference database used. The variability in taxonomic conclusions can be highlighted by the differences produced by using distinct reference databases. Previous aquaculture classifications mainly relied on mitochondrial d-loop region comparisons, which had limited identification capabilities [25]. For microorganisms, using 16S rRNA or whole-genome approaches is often more precise. Recent studies have established species boundaries using average nucleotide identity (ANI) thresholds [26].
In our research, we identified functional sequences in the Klebsiella genome, highlighting its dynamic nature and adaptability. We also emphasized the crucial role of HGT in evolution. Chen et al. [27] study on mycobacterium isolates supports this, and Dong et al. [28] stressed the importance of genetic exchange for bacterial adaptation and diversification. These findings show the inherent genomic adaptability and shared evolutionary pathways among bacterial species, prompting a re-evaluation of species demarcation criteria, taxonomic classification, and the impact of conserved sequence regions on functional traits. Al-Mustapha et al. [29] study indicates that up to 47% of E. coli genes can be classified as “auxiliary genes,” which may aid in host adaptation and are often located on MGEs. The inconsistent presence of MGEs in Klebsiella genomes from different vertebrate hosts may suggest recent host transitions. Understanding gene sequence distribution and dynamics is crucial for assessing risks related to antibiotic resistance, virulence, and environmental adaptability; further research is needed to decipher the importance and functional impacts of different gene sequences in bacterial communities, as well as their effects on ecosystems and public health [30, 31]. Our analytic results resemble the antibiotic susceptibility gene profiles of some previously reported bacterial isolates, highlighting the diversity of resistance mechanisms, including target alteration, protection, inactivation, and enhanced efflux. This layered resistance complicates infection treatment, necessitating a deeper understanding of genetic mechanisms to develop effective therapeutic strategies.
The identification of virulence factors is central to evaluating bacterial pathogenicity, as these elements are crucial for bacterial infection and clinical manifestation [32, 33]. Genome sequence analysis of the isolate revealed numerous virulence factors, including ebpA, ebpB, ebpC, srtC, ecbA, and efaA, which is key to host compliance. ebpA, a bacterial transferase, is vital for adaptation to environmental stress, especially under low-oxygen conditions [34]. Virulence-related genes, such as those for MCE and YRBE, were also detected in the isolate’s genome, underscoring their broad role in the virulence mechanisms of these bacteria; these genes are crucial for bacterial survival under low-oxygen conditions [35]. The isolate has a transferase that detoxifies reactive oxygen species, enhancing bacterial resistance to oxidative stress [36]. The identified virulence factors enable these bacteria to resist environmental stressors and effectively modulate or evade host immune defenses. Klebsiella appears to have acquired novel virulence genes from distantly related environmental bacteria, including Klebsiella sp., Neisseria sp., Clostridium sp., and Streptococcus sp. via HGT [37]. These genes include the T6SS-II (CLPB) system and KATA, GroEL, and RMLB genes. HGT-acquired genes grant Klebsiella significant functional advantages. For example, the T6SS-II system is involved in bacterial pathogenesis and host-microbe interactions, potentially increasing virulence [38]. Similarly, KATA and GroEL genes may enhance bacterial oxidative stress responses and protein folding capabilities, aiding environmental adaptation [39]. The isolate’s genome sequence contains an SDR family protein with transport functions, indicating that HGT-mediated properties enhance Klebsiella’s environmental adaptability.
Phylogenetic analysis indicates that the mr-1S strain is more closely related to Klebsiella quasipneumoniae strains. This may be due to the proximity of their breeding environments and hosts, as the Sudanese clustered isolates originate from the same location (Khartoum) as our sample collection. K. quasipneumoniae, similar to our isolate, possesses aerobactin (iutA), high viscosity, colicin V, and lacks the rmpA/rmpA2 genes, and is associated with necrotizing soft tissue infections [40]. Furthermore, genomic analysis of the Klebsiella mr-1S strain and K. quasipneumoniae reveals a significant number of LSYR family transcriptional regulators in genomic regions containing virulence and antimicrobial resistance gene clusters. It has been reported that certain LSYR family transcriptional regulators are highly expressed in crustacean aquaculture, which may be one of the reasons for their similarity [41].
A wide range of antibiotics, including β-lactams (amoxicillin, ampicillin), tetracyclines (chlortetracycline, tetracycline, hygromycin), aminoglycosides (fosfomycin, mesotryptophane), macrolides (erythromycin), sulfonamides (e.g., methotrexate combined with sulfonamides), and quinolones (oxolinic acid, flumequine, enrofloxacin), are legally approved for therapeutic and prophylactic use in aquaculture. A comprehensive review of antimicrobial resistance in China’s aquaculture industry demonstrates widespread resistance to β-lactams, tetracyclines, erythromycin, and methotrexate, with general susceptibility to chloramphenicol, tobramycin, and flumequine [42]. Notably, multidrug-resistant strains have been documented in Klebsiella, underscoring the complexity and urgency of this issue. Previous studies have identified β-lactamases in Klebsiella, but detailed information remains scarce [43]. In our research, we identified a chromosomal AmpC enzyme, classified as CARB-19, in the Klebsiella mr-1S strain, which confers resistance to various β-lactam antibiotics, including penicillins, first and second-generation cephalosporins, aminopenicillins, and carboxypenicillins. The proximity of the pqiAB gene to blaAmpC and the cusAB gene to the β-lactamase gene in our Klebsiella mr-1S strain suggests a potential link between heavy metal resistance determinants and the induction of the β-lactam resistance phenotype in these marine bacteria [44], highlighting the complex interplay of resistance mechanisms and resistance genes in the genomic landscape of Klebsiella.
Fluoroquinolones are commonly used in aquaculture. In our study, several peptide proteins were identified in the analyzed strains, with encoding genes typically plasmid-borne but found integrated into bacterial chromosomes, as reported by Weisberg [45]. Plasmid-associated antigenic determinants can spread among different bacterial species. These determinants often confer only moderate resistance to quinolones, not exceeding the clinical breakpoint alone, but can contribute to selecting additional resistance mechanisms [46]. Notably, strains with QnrA3 protein were traced back to shrimp farming, and the qnrVC6 gene, commonly associated with Vibrio cholerae, was detected. Despite expectations, all strains exhibited phenotypic susceptibility to ciprofloxacin and nalidixic acid, underscoring the complexity of resistance mechanisms and the need for a nuanced understanding of genetic elements’ interactions. ISs, with core elements like the integrase gene (intI), Int-associated recombination site (attI), and promoter, are crucial in bacterial genome evolution. Class 1 Ins play a significant role in antibiotic resistance gene dissemination among pathogens and symbiotic organisms [47, 48].
In our current study, we identified a Class 1 Ins within the genome of the Klebsiella mr-1S strain. The acquisition of such an IS has been linked to the emergence of multidrug resistance (MDR) phenotypes in Gram-negative enteric bacteria [28, 49]. Our findings corroborate this association, as the Klebsiella mr-1S strain exhibited resistance to four distinct antibiotics. This underscores the role of Class 1 Ins in the development of resistance mechanisms and highlights the importance of understanding the genetic underpinnings of MDR in clinical and environmental bacterial isolates.
Multidrug efflux or transporter protein-related genes were identified in the Klebsiella mr-1S strain genome, such as the RND family membrane fusion protein multidrug efflux pump (acrA), multidrug efflux RND transporter protein permease subunit AcrB (acrB), multidrug efflux RND transporter protein permease AcrD (acrD), efflux RND transporter protein periplasmic articulator subunit (acrE), efflux RND transporter protein permease subunit (acrF), multidrug ABC transporter protein permease/ATP-binding protein (yojl), lipid A ABC transporter protein ATP-binding protein/permease MsbA (msbA), MFS transporter protein (mdfA), phosphatidylglycolamine transferase EptA (mptA), cAMP activated global transcriptional regulator CRP (crp), exocytotic RND transporter protein periplasmic articulator subunit (oqxA), multidrug exocytotic RND transporter protein permease subunit OqxB (oqxB), multidrug exocytotic MFS transporter protein permease subunit EmrB (emrB), multidrug exocytotic MFS transporter protein periplasmic articulator subunit EmrA (emrA) and transcriptional blocker MprA (emrR). The Klebsiella mr-1S strain harbors the aadA, ebr, and dfrA12 genetic loci, which respectively encode the AadA family aminoglycoside 3′-O-nucleotidyltransferase, the SMR transporter protein for quaternary ammonium compound efflux, and the mepanipyridine-resistant dihydrofolate reductase (DfrA12). These genes confer resistance to aminoglycosides, cephalosporins, and mepanipyridine, as indicated by the respective references [50–52]. It is noteworthy that the Klebsiella mr-1S strain possesses an elevated count of such resistance genes (n = 78), implying a heightened risk of antibiotic exposure for its hosts.
In summary, NGS is a potent tool for identifying virulence determinants, with its efficacy contingent upon the currency of reference databases. It is important to note that discrepancies can arise between genotypic predictions and phenotypic expressions, particularly observed in the case of certain antibiotic classes, such as phosphomycins and quinolones. Consequently, NGS-derived data can be instrumental in signaling potential microbial resistance. While transcriptomic and proteomic assessments offer a more direct measure of resistance gene expression, WGS provides insights into the regulatory elements and environmental context influencing gene transcription. In this study, we have focused on the genomic architecture of the Klebsiella mr-1S strain to elucidate the mechanisms of resistance gene activity within aquatic microbial ecosystems and to assess the potential for genetic exchange.
The identification of prophages, CRISPR-Cas systems, and IS has shed light on the key agents that may facilitate the dissemination of virulence factors in aquatic settings. Collectively, the findings of this investigation have not only augmented the genomic repository for the Klebsiella mr-1S strain but also filled in the gaps pertaining to the Klebsiella genome. Furthermore, this study marks the inaugural demonstration of the environmental adaptability and genomic plasticity of the Klebsiella mr-1S strain.
Ethics Statement
The authors have nothing to report.
Consent
The authors have nothing to report.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Yang Xu: research, data collation, and writing–prepare the first draft. Ying Zhao: data analysis and writing preparing the first draft. Haihua Cheng: writing–preparing the first draft. Fei Peng: discussion and supervision. Qiang Gao: funding acquisition, conceptualization, and writing–review and editing. All authors have read and agreed to publish manuscript versions. Yang Xu and Ying Zhao contributed to this work equally.
Funding
This study was funded by the Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069-4), the Huzhou Special Project for “Rural Revitalization” (2021ZD2036), and China Agriculture Research System of MOF and MARA (CARS-48).
Acknowledgments
The authors have nothing to report.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




