Transcriptome Analysis Revealed the Affecting Mechanism of Two Diets, Trash Fish or Compound Feed, on Flesh Quality of Largemouth Bass (Micropterus salmoides)
Abstract
This study revealed the affecting mechanism of trash fish (TF) and compound feed (CF) on the flesh quality of largemouth bass (Micropterus salmoides) based on muscle transcriptome. Largemouth bass weighing 75.0 ± 0.1 g were given TF or CF for a period of 12 weeks. The CF group presented significantly higher feed efficiency (FE) than the TF group (p < 0.05), while there was no significant difference in specific growth rate (SGR) between the two groups (p > 0.05). A total of 604 differentially expressed genes (DEGs) meeting the significance criteria of p-value < 0.05 and |log2foldchange| > 1 were identified in the muscle transcriptome analysis. Compared to the TF group, 145 DEGs were downregulated, and 459 DEGs were upregulated in the CF group. Enrichment analysis of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways revealed significant enrichment of 401 GO terms and 13 pathways, respectively. In the CF group, there was a notable increase in gene expression in pathways such as arachidonic acid (AA) metabolism, protein processing in the endoplasmic reticulum, cholesterol metabolism, MAPK, and focal adhesion, while there was a decrease in pathways like purine metabolism, apoptosis, glycolysis/gluconeogenesis, and PPAR signaling. Real-time fluorescence quantitative PCR results indicated decreased expression of tni (troponin I, fast skeletal muscle-like) and increased expression of stni (troponin I, slow skeletal muscle-like), ftni (troponin I, fast skeletal muscle), mustn1b (musculoskeletal, embryonic nuclear protein 1b), actn2b (alpha-actinin−2), and hspb1 (heat shock protein [HSP] beta 1) in the CF group compared to the TF group. Overall, according to transcriptomics, replacing TF with CF altered the gene expression related to meat and the associated signaling pathways, leading to the meat quality improvement for largemouth bass.
1. Introduction
Trash fish (TF) is a traditional diet for carnivorous fish in aquaculture. With the progress of feed formulation and feed processing, compound feed (CF) has gradually replaced TF for carnivorous fish aquaculture. Compared with TF, CF can provide more balanced nutrition, reduce resource waste and environmental pollution, and better control the growth of fish [1].
The flesh quality comparisons of aquatic animals fed by TF and fed by compound diets have been reported in Chinese perch (Sinipercachuatsi) [2], snakehead (Ophiocephalus argus Cantor) [3, 4], large yellow croaker (Larimichthys crocea) [5, 6], Chinese mitten crab (Eriocheir sinensis) [7]. Chen et al. [2] found that the Chinese perch-fed compound diet had better growth and higher muscle hardness, brittleness, and chewing than the fish-fed baitfish. Zhang et al. [4] conducted an analysis of the proximate composition, essential amino acids, and fatty acid contents in muscle, revealing that snakehead-fed CF had higher nutritional value compared to those fed TF. In the large yellow croaker, the CF group presented better body shape, body color, meat quality, and flavor than the TF group [6]. Xiong et al. [7] evaluated the meat taste of Chinese mitten crab-fed different diets and found that crabs fed CF presented less fishy flavor and more robust taste compared to those fed TF.
Largemouth bass, Micropterus salmoides, is a member of the Perciformes order, Centrarchidae family, and Micropterus genus. Since its introduction to China from North America last century, this fish has been well received and has become an important economic aquaculture fish for its strong adaptability, fast growth, and delicious flavor without intermuscular bones. As a carnivorous fish, the aquaculture of largemouth bass depended on TF in the early stages. At present, some farmers still use TF to feed largemouth bass for some time when the TF is cheap. To date, the comparative study on TF-fed and compound diet-fed largemouth bass mainly focused on growth performance [8], gut microbiota [9], whole fish and muscle composition [10], liver and intestinal histology [11, 12], and digestive enzymes [13, 14]. Li et al. [5, 15] once reported that largemouth bass-fed TF had higher protein quality and better amino acid composition than the fish-fed compound diet, although the linoleic acid content was notably lower. Shao et al. [16] found that the TF group had lower muscle hardness, mastication, and high density of myofibrillar fibers, while the CF group showed high feed utilization. In our previous study [17], the flesh polyunsaturated fatty acids (PUFAs), n-3 PUFAs, and n-6 PUFAs levels and flesh hardness of the CF group were notably elevated, while the flesh-free amino acids level was lower than TF group. The findings of the above studies were not consistent, which may be related to the nutritional composition of TF, CF, and the size of largemouth bass.
Transcriptome sequencing technology and bioinformatics analysis have been utilized to extract data from numerous transcriptomes, unveiling gene expression patterns, regulatory pathways, and associated biological processes (BPs). Transcriptomics analysis has been widely used to study growth and development [18], nutritional regulation [19], stress response [20], and immunity [21] of aquatic animals. Moreover, this technology could help researchers deeply understand the molecular mechanism of fish muscle growth, development, and quality regulation at the gene expression level by identifying differences in gene expression related to muscle growth, metabolism, and quality under specific conditions. Wang et al. [22] once reported that the expression levels of lipid metabolism-related genes of grass carp were decreased by a high-protein diet, indicating that a high-protein diet may affect muscle quality by changing the energy metabolism pathway. Lin [23] conducted RNA sequencing on zebrafish muscles cultured at various temperatures and identified some genes associated with muscle quality regulation through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, including myf6, myog, arg2, gatm, col1a1a, and col1a2 genes.
Previously, we have examined the flesh quality of largemouth bass fed TF versus compound diet, but there remains a need for transcriptomics analysis to gain a deeper understanding of the underlying mechanisms. In this research, the transcriptome technology was utilized to examine the muscle tissue of largemouth bass that were fed either TF or CF, resulting in the identification of differentially expressed genes (DEGs). Through identifying and examining the crucial genes and metabolic pathways, the study compared and analyzed the impacts of TF and CF on the muscle quality of largemouth bass. The results will aid in comprehending the development of flesh texture and enhancing the flesh quality of largemouth bass.
2. Materials and Methods
2.1. Animal Experiments and Sample Collection
Largemouth bass were fed using two different diets: TF and CF. The TF utilized in the current experiment was ious fish (Hemiculter leucisculus) with crude protein, crude lipid, ash, and moisture levels of 172.2, 46.8, 39.2, and 741.5 g/kg, respectively. The CF was prepared by our laboratory with crude protein, crude lipid, ash, and moisture levels of 503.5, 124.3, 94.1, and 80.0 g/kg, respectively. The CF consists of 400 g/kg fish meal, 50 g/kg meat and bone meal, 80 g/kg soybean protein concentrate, 80 g/kg corn protein powder, 60 g/kg gluten powder, and 40 g/kg squid paste. The CF formula is mentioned in the explanation provided by Xu et al. [17].
Ninety fish with an average weight of 75.0 ± 0.1 g were chosen and distributed randomly into six polypropylene containers (650 L, measuring 1.0 m × 0.8 m) with 15 fish in each container. Fish were fed twice daily (7:30 AM and 4:30 PM) with the adjusted feed intake based on the fish’s feeding response to prevent waste Xu et al. [17].
Following a 12-week feeding period, all fish were deprived of food for 24 h and then sedated using 30 mg/L of MS-222. The number and weight of fish in the barrel were tallied to determine the final average weight (FBW), specific growth rate (SGR), and feed efficiency (FE). Three fish were chosen from each barrel for the sampling. The white muscle above the lateral line on the left side was collected and placed in a tube without enzymes, then promptly frozen in liquid nitrogen, and subsequently moved to −80°C for transcriptome examination.
2.2. Growth Indicators
The FBW, initial average weight, feeding period (days), and feed intake are denoted as Wt, W0, D, and FI, respectively.
2.3. RNA Extraction and Library Construction, Sequencing
Total RNA was extracted from muscle samples using TRIzol reagent (Magen). The extracted RNA was analyzed for absorbance ratios using Nanodrop ND-2000 (Thermo Scientific, USA) and for RIN values using Agilent Bioanalyzer 4150 (Agilent Technologies, CA, USA). Magnetic beads with Oligo (dT) were used to enrich the qualified mRNA samples, followed by random interruption of mRNA through the addition of fragmentation buffer. The initial cDNA strand was created by using mRNA as a template along with random hexamers, followed by the synthesis of the second cDNA strand through the addition of buffer, dNTPs, and DNA polymerase I. Finally, the double-stranded cDNA was purified using AMPure XP beads. The clean double-stranded cDNA underwent end-repair, A-tailing, and ligation with the sequencing adapter, followed by fragment size selection using AMPure XP beads. Ultimately, PCR amplification was carried out to achieve the ultimate cDNA library. The library’s quality was confirmed by detecting the size of the insert and its effective concentration. Following that, the competent library underwent sequencing with Illumina NovaSeq 6000 (or MGISEQ-T7) PE150.
2.4. Bioinformatics Analysis
2.4.1. Quality Assurance
To ensure the quality of the subsequent library analysis, it is necessary to remove the splice sequences in the original sequences (Raw Reads) and filter out the low-quality reads (Low Quality) with a base mass value ≤ 25. The elimination of substandard base sequence (representing 60% of all reads) and the exclusion of uncertain base N reads (representing more than 5%) resulted in obtaining high-quality reads suitable for further analysis.
2.4.2. Comparison of Reference Genes
The HISAT2 program was utilized to align the high-quality reads with the largemouth bass reference genome from the company to generate mapped reads for further analysis.
2.4.3. Expression Analysis
The featureCounts software was utilized to calculate the FPKM value, representing the number of transcripts per kilobase per million comparative fragments for each sample’s gene expression. The measured gene expression (FPKM value) is suitable for directly comparing gene expression variations among various samples.
2.4.4. DEGs Screening
DESeq2 was utilized for examining the variation in expression levels of FPKM values for individual genes across different samples. Padj less than 0.05 and | log2foldchange | greater than 1 were considered to be significantly different.
2.4.5. GO Enrichment Analysis
GO is a standardized functional classification system developed and maintained by the International Gene Ontology Consortium. The GO website categorizes genes and gene products in organisms into three groups: BP, molecular function (MF), and cellular component (CC).
All DEGs were compared with the known reference genomes of largemouth bass for GO functional annotation. Each DEG was associated with a GO term and subjected to Fisher’s Exact Test. p Value < 0.05 was marked as a significant difference.
2.4.6. KEGG Enrichment Analysis
The KEGG is a popular database for studying signal pathways, providing detailed information on metabolic pathways and their connections using specific graphical representations. KEGG enrichment analysis focuses on KEGG pathways and uses the corresponding reference genome of largemouth bass as background. Using Fisher’s exact test, the enrichment significance level for each pathway (p < 0.05) is determined, followed by the identification of significantly impacted metabolic and signaling pathways.
2.5. Validation of qPCR
Table 1 displays the significantly upregulated differential genes, including actn2b (alpha-actinin-2), hspb1 (heat shock protein beta 1), stni (troponin I, slow skeletal muscle-like), ftni (troponin I, fast skeletal muscle), and mustn1b (musculoskeletal, embryonic nuclear protein 1b), along with the significantly downregulated differential gene, tni (troponin I, fast skeletal muscle-like). These genes were selected for q-PCR verification analysis to confirm the reliability of RNA-seq results. cDNA was synthesized using RNAiso Plus and PrimeScript TM RT kits (Takara, Dalian, China).
| Abbreviations | Primer sequence (5′−3′) | GenBank |
|---|---|---|
| β-Actin |
|
AF300705.2 |
| tni |
|
XP_008309885.1 |
| stni |
|
XP_026217045.1 |
| ftni |
|
XP_010743757.1 |
| actn2b |
|
XP_028279163.1 |
| hspb1 |
|
XP_008288766.1 |
| mustn1b |
|
KPP66972.1 |
- Note: β-Actin is the reference gene, tni: gene-LOC119893218 (troponin I, fast skeletal muscle-like); stni: gene-LOC119918519 (troponin I, slow skeletal muscle-like); ftni: gene-LOC119893215 (troponin I, fast skeletal muscle); actn2b: alpha-actinin-2; hspb1: heat shock protein; mustn1b: musculoskeletal, embryonic nuclear protein 1b.
Primers were designed using Primer software and synthesized by Suzhou Jinweizhi Biotechnology Co., Ltd. based on the sample sequence obtained through high-throughput RNA extraction sequencing. The qPCR reaction process included an initial predenaturation at 95°C for 3 min, followed by denaturation at 95°C for 10 s, annealing at 58°C for 30 s, extension at 72°C for 30 s, and the cycle was conducted for 40 times. The target gene expression was determined using the 2−ΔΔCt method after conducting three repetitions for each sample.
3. Results
3.1. Growth Performance
After 12 weeks of feeding, all the fish presented good growth performance without mortality. The FE was notably greater in the CF group compared to the TF fish group (p < 0.05), while there were no significant disparities in survival rate and SGR between the two groups (p > 0.05) (Table 2).
| Parameters | TF | CF |
|---|---|---|
| IBW/g | 74.9 ± 0.2 | 75.1 ± 0.2 |
| FBW/g | 271.2 ± 16.1 | 266.3 ± 2.6 |
| SGR (%/day) | 1.53 ± 0.06 | 1.51 ± 0.02 |
| FE | 0.92 ± 0.04b (0.24 ± 0.01)∗ | 1.13 ± 0.02a |
| SR% | 100 | 100 |
- Note: Values are mean ± SD (n = 3) of three replicates, and different superscripts in the same row indicate significant differences (p <0.05).
- Abbreviations: CF, compound feed; FBW, final average weight; SGR, specific growth rate; TF, trash fish.
- ∗Feed efficiency in parentheses is calculated based on fresh weight.
3.2. Raw Data, De Novo Assembly, and Annotation and Reference Gene Alignment
In Table 3, a total of 360,185,972 raw reads and 360,033,330 clean sequence reads were obtained after filtering. The Q20, Q30, and GC content all had a high quality with values exceeding 97%, 93%, and 51.55%−52.29%, respectively, suggesting good overall base quality.
| Sample | Raw_reads | Clean_reads | Clean_bases | Error (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| TF-1 | 40,421,692 | 40,405,882 | 5.9G | 0.04 | 97.63 | 93.37 | 51.88 |
| TF-2 | 68,357,756 | 68,330,462 | 9.97G | 0.04 | 97.79 | 93.78 | 52.29 |
| TF-3 | 61,210,452 | 61,186,936 | 8.86G | 0.04 | 98.08 | 94.5 | 52.03 |
| CF-1 | 47,968,148 | 47,943,130 | 7.02G | 0.04 | 98.09 | 94.6 | 51.55 |
| CF-2 | 54,698,838 | 54,679,550 | 8G | 0.04 | 97.95 | 94.17 | 51.93 |
| CF-3 | 87,529,086 | 87,487,370 | 12.81G | 0.04 | 98.06 | 94.56 | 51.98 |
- Note: (1) Sample: sample name. (2) Raw reads: the total number of entries of the original sequencing data. (3) Clean reads: the total number of items of sequencing data after quality control. (4) Clean bases: the total amount of sequencing data after quality control. (5) Error rate (%): the average error rate of sequencing bases corresponding to quality control data is generally below 0.1%. (6) Q20 (%) and Q30 (%): Evaluate the quality of sequencing data after quality control. Q20 and Q30 refer to the percentage of bases with sequencing quality above 99% and 99.9%, respectively; generally, Q20 is above 85% and Q30 is above 80%. (7) GC content (%): the percentage of the total number of G and C bases corresponding to the quality control data.
- Abbreviations: CF, compound feed; TF, trash fish.
Comparing the above clean sequences with the reference genebank, the sequenced sequences that could be compared to the genome for each sample were ≥93.02% (TF-1), which exceeded the standard of 70% (Table 4). Such results showed that the quality of sequencing data was good, and they could be used for subsequent analysis.
| Sample name | Total reads | Total mapped | Multiple mapped | Unique mapped | Nonsplice reads | Splice reads |
|---|---|---|---|---|---|---|
| TF-1 | 40,405,882 | 37,586,039 (93.02%) | 7,649,054 (18.93%) | 29,936,985 (74.09%) | 10,412,467 (25.77%) | 19,524,518 (48.32%) |
| TF-2 | 68,330,462 | 63,940,177 (93.57%) | 12,735,391 (18.64%) | 51,204,786 (74.94%) | 17,491,473 (25.60%) | 33,713,313 (49.34%) |
| TF-3 | 61,186,936 | 57,520,121 (94.01%) | 10,547,000 (17.24%) | 46,973,121 (76.77%) | 16,796,115 (27.45%) | 30,177,006 (49.32%) |
| CF-1 | 47,943,130 | 45,564,025 (95.04%) | 8,938,682 (18.64%) | 36,625,343 (76.39%) | 13,894,598 (28.98%) | 22,730,745 (47.41%) |
| CF-2 | 54,679,550 | 51,878,393 (94.88%) | 10,389,301 (19.00%) | 41,489,092 (75.88%) | 14,566,043 (26.64%) | 26,923,049 (49.24%) |
| CF-3 | 87,487,370 | 82,951,177 (94.82%) | 16,348,159 (18.69%) | 66,603,018 (76.13%) | 23,338,776 (26.68%) | 43,264,242 (49.45%) |
- Note: (1) Sample: sample name; (2) Total reads: the statistics of the number of filtered sequencing sequences (i.e., clean reads); (3) Total mapped: the number of clean reads that can be located on the genome; (4) Multiple mapped: the number of clean reads with multiple alignment positions on the reference sequence; (5) Unique mapped: the number of clean reads with unique alignment positions on the reference sequence; (6) Nonsplice reads: the number and proportion of the whole reads aligned to the reference genome; (7) Splice reads: total mapped, the statistics of sequencing sequences (also known as Junction reads) aligned to two exons.
- Abbreviations: CF, compound feed; TF, trash fish.
3.3. DEGs Analysis
The analysis of DEGs was conducted using DESeq2, with the results displayed in Figure 1. The comparison of TF and CF groups presented a total of 604 DEGs (p < 0.05). Among them, 459 genes were significantly upregulated, as shown by the red scatterplot, while 145 genes were significantly downregulated, as shown by the blue scatterplot.

3.4. DEGs Clustering
Gene cluster analysis was conducted by comparing the gene expression levels in each sample to visually represent the expression patterns of the 604 DEGs across different samples (Figure 2).
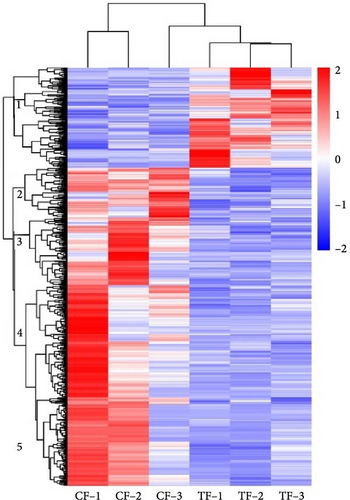
As shown in Figure 2, all downregulated DEGs are clustered into branch 1, such as f13a1b (coagulation factor xiii a chain b), adpgk (ADP dependent glucokinase), wnt7b (wnt family member 7b), and nudt2 (nudix hydrolase 2). The unique genes have regulatory functions in blood clotting, sugar processing, cell growth and specialization, and nucleic acid metabolism.
atcaya (atcay kinesin light chain interacting caytaxin), amotl2a (angiomotin like 2a), and vegfc (vascular endothelial growth factor c) have similar branches and cluster into branch 2. They play regulatory roles in neural tissue development and angiogenesis.
smoothelin like 2 (smtn), rna binding motif protein 22 gene (rbm22), and insulin-like growth factor binding protein 5a (igfbp5a) present comparable expression profiles and cluster into branch 3. They have important functions in organizing the actin cytoskeleton and controlling vascular contraction, as well as in activating transcription factors, regulating RNA metabolism, controlling gene expression, and influencing cell growth, differentiation, and apoptosis.
pparg (peroxisome proliferator-activated receptorγ), ptgdsb.1 (prostaglandin d2 synthase b, tandem duplicate 1), lpl (lipoprotein lipase), nceh1a (neutral cholesterol ester hydrolase 1a); ppp1r1b (protein phosphatase 1 regulatory inhibitor subunit 1b), ppp1r3g (protein phosphatase 1 regulatory subunit 3g), c7b (complement c7b), cfd (complement factor d) are grouped in category 4, playing roles in lipid metabolism, glucose metabolism, immunity, and more.
actn2b (actinin alpha 2b), myoz2b (myozenin 2b), tpm2 (tropomyosin 2), tpm3 (tropomyosin 3), mustn1a (musculoskeletal, embryonic nuclear protein 1a), mybpc1 (myosin binding protein c1), and heat shock protein (HSP) family members hspb1, hspb7, hspa8b, are grouped in branch 5, where they have significant regulatory functions in muscle growth and development.
3.5. GO Functional Annotation and Enrichment Analysis
GO enrichment analysis indicated that 604 DEGs were enriched into BP, MF, and CC. A total of 401 entries were selected, and the top 10 significant functional items were selected and represented by a bar chart (Figure 3).
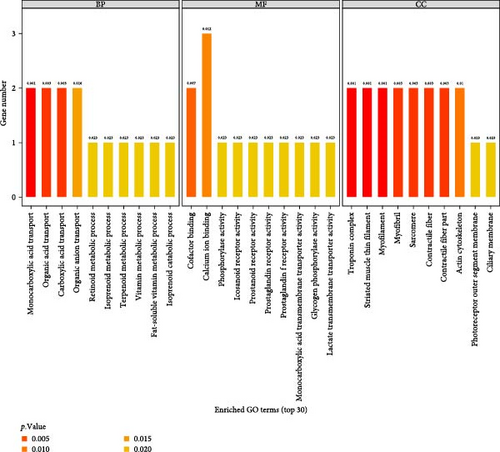
DEGS are predominantly found in transport and metabolic processes in the BP classification, such as monocarboxylic acid transport, organic acid transport, carboxylic acid transport, organic anion transport, retinoid metabolic process, isoprenoid metabolic process, terpenoid metabolic process, vitamin metabolic process, fat-soluble vitamin metabolic process, and isoprenoid catabolic process. The above BPs are closely related to organic acid transport, lipid metabolism, and vitamin metabolism.
DEGs in the MF classification participate in various functions such as cofactor binding, calcium ion binding, phosphorylase activity, icosanoid receptor activity, prostanoid receptor activity, prostaglandin receptor activity, prostaglandin F receptor activity, monocarboxylic acid transmembrane transporter activity, glycogen phosphorylase activity, and lactate transmembrane transporter activity. The functions include cofactor binding, calcium ion binding, phosphorylase activity, icosanoid receptor activity, prostanoid receptor activity, prostaglandin receptor activity, prostaglandin F receptor activity, monocarboxylic acid transmembrane transporter activity, glycogen phosphorylase activity, and lactate transmembrane transporter activity.
The troponin complex, striated muscle thin filament, myofilament, myofibril, sarcomere, contractile fiber, actin cytoskeleton, photoreceptor outer segment membrane, and ciliary membrane are the primary components enriched in the CC classification. Cell components are essential for structural organization, functional regulation, and muscle contraction, achieved by interacting with protein complexes, transmitting signals, and connecting with other organelles.
3.6. Analysis of Functional Annotation and Enrichment in the KEGG Database Was Performed
According to KEGG analysis, the signal transduction category in the environmental information processing class had the highest number of annotated genes, followed by the endocrine system, immune system, and circulatory system in the organismal systems category. Next, they were the cell processes category, encompassing cell community—eukaryotes, transport, and catabolism, along with metabolic pathways and the digestive system in the organismal systems category. In addition, a few DEGs were enriched in other functional categories.
The findings indicated that DEGs were concentrated in 98 pathways. Among them, the top 20 pathways were selected using the p-value and visualized in a scatterplot (Figure 4). Figure 4 displays that the estrogen signaling pathway had the most enriched DEGs (seven DEGs), then the MAPK signaling pathway and adrenergic signaling in cardiomyocytes (six DEGs). The cardiac muscle contraction pathway enriched five DEGs, and four DEGs were enriched in the antigen processing and presentation pathway, longevity regulating pathway—multiple species, spliceosome, protein processing in the endoplasmic reticulum, and endocytosis. The arachidonic acid (AA) metabolism, focal adhesion, renin secretion, and oxytocin signaling pathway enriched three DEGs, respectively. In addition, 10 DEGs were enriched in cholesterol metabolism, adherens junction, carbon metabolism, cortisol synthesis and secretion, and vitamin digestion and absorption with two DEGs per pathway.
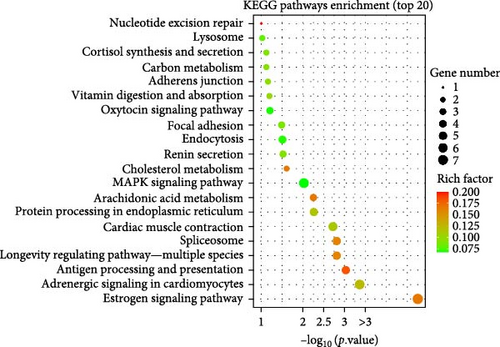
Table 5 displays 13 pathways chosen from the top 20 significantly enriched pathways. In comparison to the TF group, the DEGs in the CF group showed notable enrichment in pathways such as estrogen signaling, adrenergic signaling in cardiomyocytes, antigen processing and presentation, longevity regulating pathway-multiple species, spliceosome, cardiac muscle contraction, AA metabolism, proteins processing in endoplasmic reticulum, MAPK signaling, cholesterol metabolism, endocytosis, focal adhesion, and renin secretion.
| ID | Description | Test | Ref | p-Value | FDR |
|---|---|---|---|---|---|
| ko04915 | Estrogen signaling pathway | 7 | 42 | 1.75e−05 | 1.71e−03 |
| ko04261 | Adrenergic signaling in cardiomyocytes | 6 | 49 | 4.33e−04 | 2.12e−02 |
| ko04612 | Antigen processing and presentation | 4 | 22 | 9.40e−04 | 3.04e−02 |
| ko04213 | Longevity regulating pathway—multiple species | 4 | 25 | 1.55e−03 | 3.04e−02 |
| ko03040 | Spliceosome | 4 | 25 | 1.55e−03 | 3.04e−02 |
| ko04260 | Cardiac muscle contraction | 5 | 44 | 0.001916 | 0.031301 |
| ko00590 | Arachidonic acid metabolism | 3 | 18 | 0.005642 | 0.06911 |
| ko04141 | Protein processing in endoplasmic reticulum | 4 | 35 | 0.005506 | 0.06911 |
| ko04010 | MAPK signaling pathway | 6 | 89 | 0.009543 | 0.103915 |
| ko04979 | Cholesterol metabolism | 2 | 12 | 0.0248 | 0.243045 |
| ko04144 | Endocytosis | 4 | 58 | 0.031404 | 0.246841 |
| ko04510 | Focal adhesion | 3 | 34 | 0.032744 | 0.246841 |
| ko04924 | Renin secretion | 3 | 33 | 0.030292 | 0.246841 |
Among the 13 pathways, four pathways were chosen AA metabolism, protein processing in endoplasmic reticulum, cholesterol metabolism, as well as MAPK signaling pathway and focal adhesion. They were related to nutrition metabolism and meat quality, and DEGs in these pathways were significantly upregulated (p < 0.05). In addition, some signaling pathways related to muscle quality were also screened from other nonsignificantly enriched pathways (p > 0.05) with downregulated DEGs, such as glycolysis/gluconeogenesis, apoptosis, PPAR signaling pathway, and purine metabolism. Although these pathways were not significantly enriched in the differential gene analysis, they still showed varying degrees of DEG enrichment, indicating that they may have some regulatory effects on muscle quality.
3.7. qPCR Result
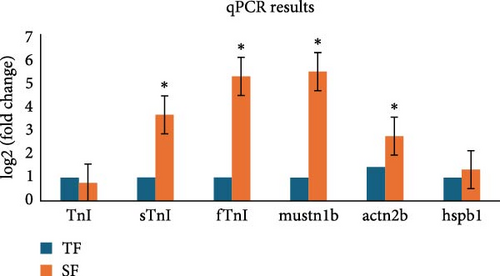
The qPCR results are shown in Figure 5. In comparison to the TF group, the CF group showed a decrease in tni expression and an increase in stni, ftni, mustn1b, actn2, and hspb1 expressions. The findings were in line with the RNA-Seq sequencing outcomes.
4. Discussion
4.1. Growth Performance
In this trial, the protein content of TF was 66.62% (dry weight), while the CF was 54.73% [24]. The SGR and survival rate show no discrepancy between the two groups, but the TF group exhibited significantly lower protein efficiency than the CF group. The findings showed that the protein in CF was sufficient for largemouth bass growth, while an excess of protein led to waste and higher feed expenses. El-Hammady et al. [25] reported that European seabass (Dicentrarchus labrax) fed on TF or pelleted feed presented similar growth performance and survival rate. Comparable findings were also discovered in Lates calcarifer and Epinephelus fuscoguttatus [26]. The above findings indicated that the production performance of carnivorous fish fed CF will not be decreased, and the FE may be improved. Thus, CF could completely replace the TF application in largemouth bass aquaculture.
4.2. Muscle Transcriptome and Related Gene Expression
Our previous study showed that largemouth bass fed with TF had higher free amino acid content in muscle, but the muscle of fish fed with CF had higher hardness, heat-insoluble collagen content, PUFA level, and lower drip loss and myofiber diameter [24]. In addition to exogenous factors such as nutrition and environment, intrinsic factors, and genetic effects may play more important roles in the forming of muscle quality [27]. The growth and development of fish muscle include the proliferation of new myofibers and the growth of existing myofibers. Therefore, identifying factors that regulate muscle growth and development can provide potential targets for improving the muscle quality of fish [28].
Bioinformatics analysis of the 604 DEGs obtained from the CF and TF groups revealed that DEGs are enriched in pathways related to muscle nutrition metabolism and quality. Among them, AA metabolism, endoplasmic reticulum protein processing, cholesterol metabolism, MAPK signaling pathway, and focal adhesion were significantly affected. Glycolysis/gluconeogenesis, apoptosis, PPAR signaling pathway, and purine metabolism were also affected to varying degrees.
4.2.1. AA Metabolism
AA is an essential PUFA with crucial roles in lipid metabolism [29, 30]. AA can engage in metabolic functions by interacting with enzymes like COX, LOX, and CYP450, resulting in the creation of substances like PG, LT, and 20-HETEs. Those metabolic products exert a regulatory effect on fat metabolism, which has been confirmed in grass carp [31]. L-PGDS, also known as prostaglandin D synthase, is a versatile protein capable of producing D series prostaglandins and serving as a carrier for lipophilic molecules [32, 33]. The current research found that the ptgdsb.1 gene (prostaglandin D2 synthase b, tandem duplicate 1) was notably increased in the CF group compared to the TF group. The ptgdsb.1 functions as a carrier protein for lipophilic compounds involved in lipid metabolism, as well as catalyzing the conversion of AA into prostaglandin D2 (PGD2). After the production, PGD2 is released from cells and acts on surrounding tissues and organs to regulate lipid metabolism [34, 35]. It was found that AA metabolic pathway genes are generally conserved in eukaryotes, and their evolutionary dynamics may be that the emergence of mitochondria in eukaryotes leads to new antioxidant defense responses [36]. Muscle antioxidant capacity is one important indicator to evaluate the meat quality characteristics of aquatic products [22]. A decline in antioxidant capacity can result in oxidative damage and the accumulation of free radicals in the body, impacting the structure of muscle cells, metabolic functions, and the development of flavor compounds, ultimately diminishing muscle quality [37]. Previous study has found that dietary changes in protein level affected the GPx gene expression in muscle of crucian carp (Carassius carassius Triploid), thereby affecting the antioxidant status of muscle [38], reducing oxidative damage, and improving muscle quality [39]. The lipid mediator 20-HETE produced by AA metabolism can induce the production of ROS and cause oxidative damage [40]. In our previous study [17], the screening of muscle metabolites showed that the 20-HETE in the CF group was decreased when compared with the TF group. Moreover, there was a notable increase in gpx1b expression in the CF group by the transcriptome results. It is suggested that gpx1b may enhance antioxidant function, inhibit the production of 20-HETE, and protect muscle cells from oxidative damage, thereby improving muscle quality and stability.
4.2.2. Endoplasmic Reticulum Protein Processing
The endoplasmic reticulum protein processing pathway is involved in protein folding, classification, and degradation. HSPs, acting as molecular chaperones, are produced in response to stress and play roles in the proper folding and stability of proteins. They can attach to irregular or unsteady proteins, aiding in their proper folding and durability, and preventing the formation of detrimental clumps [41], thus maintaining the normal structure and function of proteins in muscle. In a study conducted by Lin [23], RNA sequencing was carried out on zebrafish muscle exposed to varying temperatures. The results revealed a notable enrichment of hsp70 (heat shock proteins 70) and hsp40 (heat shock proteins 40) genes in endoplasmic reticulum protein processing, leading to alterations in protein synthesis and degradation pathways. HSPs help break down improperly folded proteins to restore balance in the endoplasmic reticulum near misfolded channels involved in endoplasmic reticulum-associated protein degradation [42]. The current findings indicated a notable increase in heat shock-related proteins within this pathway. There is speculation that HSPs impact the folding and stress resistance, protein integrity, and metabolic state of fish muscles through their involvement in regulating protein processing in the endoplasmic reticulum, ultimately influencing muscle quality.
4.2.3. Cholesterol Metabolism
Cholesterol is involved in the synthesis, decomposition and transport of fatty acids, maintaining energy balance and fat storage. Neutral cholesterol esterase 1a (Nceh1a) is an enzyme involved in cholesterol ester metabolism, which hydrolyzes cholesterol esters into free cholesterol and fatty acids [43, 44]. Cholesterol can contribute to various biological functions like the synthesis of cell membranes and hormones, and cells can also absorb and use fatty acids [44, 45]. In a prior investigation, it was discovered that the concentrations of polyunsaturated fats, specifically EPA and DHA, in the muscle tissue of the CF group exceeded those in the TF group [17]. Transcriptome results showed that nceh1a expression was markedly elevated in the CF group compared to the TF group. Maybe the CF promoted the hydrolysis of cholesterol esters, releasing more fatty acids for absorption and utilization, resulting in higher and more comprehensive levels of fatty acids in the muscle tissue.
4.2.4. MAPK Signaling Pathway
The MAPK signaling pathway is a highly conserved three-tier kinase cascade pathway for transmitting signals. MAPK activation can control various BPs like cell growth, specialization, change, and cell death through the phosphorylation of targets like transcription factors, cytoskeleton-related proteins, and enzymes. The activation of the MAPK signaling pathway can enhance the expression of heat shock factor HCF1 (heat shock factor, leading to increased binding of HCF1 to the heat shock element of HSP70 and elevated synthesis of HSP70 [46]. HSPs play important roles in muscle quality with significant effects on muscle texture, tenderness, shear force, etc. [47, 48]. Elevated expression of HSPs can impede the programed cell death of cow muscle cells, slow down the breakdown of myofibrillar proteins, and lessen muscle tenderness [48]. In Brazilian native Nelore cattle, HSP70 was found to be positively correlated with the shear force of the pectoral muscle after slaughter [47]. The increase of shear force results in greater muscle firmness; therefore, the elevated HSP70 levels can enhance muscle firmness. AP-1 is a protein responsible for controlling cell growth, development, and cell death. Gao et al. [49] reported that the elevated levels of AP-1 gene family members promoted muscle development and decreased fat levels in the muscles of pigs. Dietary broad bean has been reported to increase the DEGs related to MAPK and other pathways, thereby increasing the flesh hardness and chewiness of grass carp [50]. The current findings indicated a notable increase in the expression of hsp70b and June (transcription factor AP-1) in the MAPK signaling pathway in the CF group compared to the TF group. In previous studies, it was found that the CF group had notably greater muscle hardness and shear force than the TF group [17]. Maybe CF can regulate MAPK to inhibit cell apoptosis, delay protein degradation of myofibrillar proteins, reduce muscle fat content, and thus increase muscle hardness and shear force.
4.2.5. Focal Adhesion
Macromolecular complexes on the cytoplasmic side of focal adhesions bind transmembrane receptors like integrin to the actin cytoskeleton, transmitting signals that control cell adhesion, migration, proliferation, differentiation, and gene expression [51, 52]. Focal adhesion plays an important roles in signal transduction in various BPs, including myogenesis [53]. Genes encoding adhesion proteins are considered candidates for meat quality [54]. CHAD, a protein-rich in leucine, can enhance cell adhesion through its interaction with integrin α (2) β (1) [55]. The increased Col6a1 level can control Chad and Camk2 in intramuscular adipocytes, leading to higher lipid droplet formation and lower cell growth, ultimately impacting the deposition of IMF [56]. In addition to cell adhesion, focal adhesion can also be used as a sensor to transmit the status of the extracellular matrix (ECM) to cells, thereby affecting cell migration, proliferation, differentiation, survival, and cell morphological changes [57]. Focal adhesion may have a synergistic effect with ECM receptor interaction to affect the content of collagen, thereby affecting muscle hardness. Chad was found to be significantly increased in the focal adhesion signaling pathway in this study, potentially playing a role in controlling the actin cytoskeleton and impacting myogenesis and collagen synthesis.
4.2.6. Glycolysis/Gluconeogenesis
Glucose is degraded into pyruvate in the cytoplasm through enzymatic catalysis, simultaneously generating ATP. The change in muscle pH after slaughter glycogen content, glycolytic enzyme activity, and substrates in muscle before slaughter are all closely linked to the glycolytic potential (GP). Generally, a greater pH change means a stronger GP [58]. The value of pH is a key indicator reflecting muscle quality, and the pH change directly affects the color, tenderness, and water-holding capacity of meat [59]. ADPGK, also known as ADP-dependent glucokinase, can utilize ADP as a phosphate donor to facilitate the conversion of glucose into glucose-6-phosphate. This enzyme is essential for energy metabolism by participating in the glycolysis pathway [60]. Yu et al. [61] found that adding guanidineacetic acid (GAA) to the diet of channel catfish (Ictalurus punctatus) raised the phosphocreatine ATP levels in the muscles, decreased the utilization of muscle glycogen for energy, and ultimately lowered the lactate content. Wei et al. [62] reported that dietary Hyp supplements improved the muscle quality of large yellow croaker by enhancing the glycolytic pathway using a comprehensive analysis of transcriptomics and metabolomics. ADPGK was found to be highly concentrated in the glycolysis/gluconeogenesis pathway in this research, and the CF group displayed notably reduced adpgk levels compared to the TF group. This suggests that feeding CF alleviates anaerobic glycolysis, reducing lactate production. The relatively higher pH value leads to the contraction of myofibrils, resulting in firm, compact muscle texture and the improvement of water-holding capacity. A high capacity to hold water will assist in preserving flavor compounds [63], improving meat color [64], and thus enhancing muscle quality.
4.2.7. Apoptosis
Apoptosis, also referred to as programed cell death, is a regulated and organized cellular demise controlled by multiple genes [24]. Postmortem aging is a crucial method for enhancing the quality of meat. Apoptosis is crucial in controlling the process of postmortem aging. Therefore, by controlling the process of apoptosis, the meat parameters can be regulated to improve meat quality [24]. Studies have shown a connection between apoptosis and the tenderness, water retention, and color of meat [65]. Typically, apoptosis involves the breakdown of myofibrillar protein, a crucial element that impacts water retention ability [65] and tenderness. Positive apoptotic nuclei in grass carp were found to be positively correlated with droplet loss and negatively correlated with shear force and water-holding capacity [66]. Cramer et al. [67] reported that the inhibition of apoptosis occurrence increased the shear force and hardness of lamb lumbar muscle. Chen et al. [68] found that the degradation products of chicken myofibrillar protein treated with apoptosis inducer were increased significantly, and the tenderness was improved. Cathepsin is essential for the quality of meat, which existsin cell lysosomes. When the lysosomes are damaged, cathepsins will be released into the cytoplasm and participate in the regulation of important processes such as apoptosis [69, 70]. Li et al. [71] reported that the activity of cathepsin L and D in iced tilapia fillets was significantly correlated with hardness, adhesiveness, and chewiness. In rainbow trout, cathepsin D, B, and L also exerted important effects on flesh texture, and cathepsin D had the greatest effect on hardness [72]. In this study, gene-LOC119890600 (cathepsin D-like) was downregulated in this pathway. It can be speculated that feeding CF can reduce myofibrillar protein degradation products and increase hardness and water-holding capacity by inhibiting apoptosis.
4.2.8. PPAR Signaling Pathway
The PPAR gene family consists of three variants, PPARα, PPARβ, and PPARγ, that primarily influence adipocyte differentiation through controlling lipid metabolism gene expression. Following activation, PPAR can start the fat production process by controlling the activity of genes involved in fat synthesis, such as LPL and FAS. PPAR is a crucial signaling factor for lipid synthesis [73, 74]. The primary reasons for fat metabolism and deposition in animals are the activation of the PPAR signaling pathway and the expression alterations of regulatory genes associated with this pathway [75]. Fat can help increase the flavor, taste, and tenderness of fish meat. PPAR-α is an important part of the PPAR signaling pathway, which is involved in many pathways, such as ketogenesis, lipid transport, and fatty acid biosynthesis. Unsaturated fatty acids and other ligands activate PPAR-α, forming heterodimers that control the expression of downstream target genes like malic enzyme 1 gene (ME1) and play roles in fatty acid biosynthesis [76]. ME is one of the important enzymes for NADPH (reduced coenzyme II) production [77], and it can initiate the oxidative decarboxylation of malic acid to generate NADPH, a crucial source of NADPH in the production of fatty acids [78]. In animals, NADPH plays a crucial role as a coenzyme in the synthesis of fatty acids and the elongation of carbon chains. Yang et al. [79] found that dietary Eucommia ulmoides bark and leaf supplementation promoted the growth, lipid metabolism, and meat quality of grass carp through mTOR and PPAR signaling pathways. The supply of NADPH directly affects the synthesis speed of lipids. In this study, LOC119884012 (NADP-dependent malic enzyme) in the CF group was downregulated in this pathway. Due to the lower muscle crude fat content and higher metabolite DL malic acid level (upregulated) in the CF group than those in the TF group, it is speculated that the oxidation and decarboxylation reaction of malic acid in the CF group was hindered, leading to a reduction in NADPH and potentially impacting the synthesis of fatty acids. Consequently, the CF group exhibited greater muscle firmness than the TF group, although there may be a reduction in flavor.
4.2.9. Purine Metabolism
Volatile components of lipid metabolism, amino acids, and nucleotides are important contributors to flavor. Du et al.[80] found that purine metabolism played important roles in improving the meat quality of crucian carp through comprehensive analysis of transcriptomics and metabolomics. Inosinic acid (IMP), also known as hypoxanthine (Hx) nucleotide or Hx acid, is a flavor substance with umami enhancer [81], which can participate in the Maillard reaction and is an important precursor to produce meat flavor [82]. Thus, IMP is usually used as an important index to reflect the umami of meat [83]. During the heating process, a significant portion of ATP in fish meat will be converted to IMP, enhancing the flavor of fish meat [84]. AMP and GMP play important roles in purine metabolism. AMP serves as the precursor for IMP, which is then broken down into xanthine (Xan) and Hx [85, 86]. AMP and IMP presented an umami taste, while Hx and Xan were the sources of bitterness. nudt2 (nudix hydrolase 2) is a nucleic acid diphosphohydrolase involved in nucleotide metabolism regulation and significantly affecting the metabolism of AMP and IMP in cells. The main substrate of nudt2 is considered Ap4A (tetraphosphorylated adenosine), which can be hydrolyzed to produce AMP and ATP [87]. AMP is an important intermediate product generated from IMP synthesis, and ATP can be decomposed to produce IMP. The present findings indicated that the distinct gene nudt2 was highly concentrated in purine metabolism, with the CF group exhibiting lower levels than the TF group. It is speculated that this gene may affect the metabolism of IMP through purine metabolism, thus influencing the flavor of fish meat.
4.2.10. Expression of Genes Related to Muscle Structure and Function
Troponin I, skeletal troponin I, fast troponin I, and mustn1b are crucial for muscle growth and movement TnI, sTnI, and fTnI belong to troponin, and they are one of the myofibrillar proteins required for skeletal muscle contraction. TnI-slow (slow skeletal muscle troponin I) and TnI-fast (fast skeletal muscle troponin I) are two variations that can impact muscle fiber composition and ultimately influence the quality of meat [88]. The current findings from qPCR indicated a decrease in tni expression with an increase in stni and ftni levels in the CF group. This could be the muscle’s natural reaction to environmental changes, stress, and hormonal influences, leading to alterations in muscle fiber composition. Mustn1 is a specific musculoskeletal protein involved in cell proliferation and differentiation during skeletal muscle development and regeneration [89]. mustn1 gene expression was notably increased in the CF group, indicating that CF consumption may enhance muscle development and growth. actn2b and hspb1 are crucial for preserving the structure and functionality of muscle cells. Microfilaments formed by actn2b are a part of the eukaryotic cell skeleton and are involved in cell junction, cell shape, apoptosis, migration, proliferation, and cell signal transduction [90]. actn2b binds with several proteins associated with focal adhesions, including vinculin (VCL) [91, 92] and integrin [93, 94], to maintain muscle cell structure. Spb1 exerts important roles in stress response and protein quality control. The present findings showed that providing CF led to a notable increase in the expression levels of actn2b and hspb1, indicating that the development of the cytoskeleton was effectively sustained, thereby enhancing muscle growth and quality.
5. Conclusion
In the present study, largemouth bass fed with CF showed higher FE than the fish fed with TF, but both presented a similar SGR. The transcriptomics analysis revealed 604 differential genes with 145 downregulated genes and 459 upregulated genes in the CF group. In comparison to the TF group, the CF group showed lower expression of tni and higher expression of stni, ftni, mustn1b, actn2b, and hspb1. KEGG pathway analysis revealed significant upregulation of AA metabolism, protein processing in endoplasmic reticulum, cholesterol metabolism, MAPK, and focal adhesion signaling pathways, while apoptosis, glycolysis, PPAR, and purine metabolism were downregulated in the CF group. These changes may affect the flesh quality of largemouth bass. In the future, the key components and functions of signaling pathways will be further explored to improve the flesh quality of largemouth bass.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Min Feng: conceptualization, methodology, formal analysis, investigation, data curation, writing–original draft. Xiaoying Xu: conceptualization, investigation, methodology, data curation. Zhifen Xu: conceptualization, investigation, methodology, validation, formal analysis. Xiangjun Leng: funding acquisition, resources, writing–review and editing. Xiaoqin Li: funding acquisition, resources, writing–review and editing, supervision. The first author is Min Feng and the co-first author is Xiaoying Xu, and both authors contributed equally to this work.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFD0900200).
Open Research
Data Availability Statement
Data will be made available upon request.




