Occurrence and Antimicrobial Resistance Pattern of Listeria monocytogenes Isolated From Bovine’s Milk and Meat in Mekelle City, Ethiopia
Abstract
Listeria monocytogenes is an opportunistic and emerging foodborne zoonotic pathogen that encompasses a diversity of strains with varied virulence and can cause serious human and animal infections worldwide. However, in Mekelle City, the actual prevalence and the antimicrobial susceptibility pattern of L. monocytogenes were not done. Hence, this cross-sectional study was conducted from December 2016 to June 2017 to determine the prevalence of L. monocytogenes and its serotypes and antimicrobial resistance pattern of isolates in Mekelle City, Ethiopia. A total of 768 (n = 384 of milk and n = 384 meat) samples of bovine origin were collected using a purposive random sampling technique. Isolation and identification of L. monocytogenes were done according to standard and recommended bacteriological procedures. Polymerase chain reaction (PCR) amplification of Iap, Imo0737, ORF2819, and ORF2110 genes was performed. In vitro antimicrobial susceptibility testing was performed using agar disk diffusion method. The overall prevalence of L. monocytogenes was 3.39%. The prevalence rates of L. monocytogenes were 4.17% and 2.6% in meat and milk samples, respectively. There was a statistically significant difference (p < 0.05) in the prevalence rates of the organism in meat samples collected from abattoir (1.67%), butcher shops (8.33%), and restaurants (8.33%). Serovars that were identified belonged to 1/2b and 4b. Large proportions of isolates were highly susceptible to ampicillin (88.46%) and vancomycin (84.62%). However, the isolates had shown the highest level of resistance against nalidixic acid (96.15%). Moreover, 42.31% of the isolates developed multidrug resistance. Hence, both its occurrence and development of a multidrug resistance indicated the need to practice adequate cocking of bovine origin foods before consumption and rational use of antimicrobials both in veterinary and human treatment regimens with regular surveillance of antimicrobial resistance to combat multidrug resistance.
1. Introduction
Foodborne diseases remain a major public health problem across the globe. The problem is severe in low-income countries due to difficulties in securing optimal hygienic food handling practices and with the increase in the consumption of raw products of animal origin. Hence, food safety and particularly the safety of products of animal origin are an increasingly significant issue concerning human health [1–3]. There are over 200 known microbial, chemical, or physical agents that can cause illness when ingested [4]. Out of the agents associated with the current worldwide increase in cases of food-borne diseases, Listeria monocytogenes is implicated in numerous outbreaks of invasive listeriosis reported globally, characterized by septicemia, meningoencephalitis, and maternal–fetal infection leading to abortion [5, 6]. Listeriosis manifests itself clinically in ruminants as encephalitis, neonatal mortality (abortion), and septicemia [7].
Listeria spp. are gram-positive, psychrotrophic, facultative anaerobic, nonsporulation, and intracellular bacteria that are widely distributed in the natural environment [8, 9]. The identification of L. swaminathanii brings the total number of Listeria species to 27 [10]. However, L. monocytogenes and Listeria ivanovii are the two species within this genus known to cause infection in humans and animals, with L. monocytogenes being the leading cause of human clinical listeriosis [11].
L. monocytogenes comprises 13 serotypes with different virulence potentials, namely, 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4ab, 4b, 4c, 4d, 4e, and 7, based on somatic (O factor) and flagella (H factor) antigens [12, 13]. However, more than 95% of human listeriosis is commonly associated with isolates that belong to serotypes 1/2a, 1/2b, and 4b, and of these, serotype 4b has been related to the most recent outbreaks of listeriosis, and serotypes 1/2a and 4b are commonly reported in animals [14–20].
L. monocytogenes is a severe risk for the health of specific population groups such as the elderly, immunocompromised individual pregnant women and newborns. Listeriosis can also affect animals (mainly ruminants), and acquisition of the disease is generally due to the consumption of contaminated feed (particularly silage) or food (such as ready-to-eat) [21–24]. Despite its low incidence, listeriosis is associated with a high fatality rate [22]. The central characteristics of L. monocytogenes contributing to food-borne transmission are the ability to grow as low as −0.4°C; resist heat, salt, nitrite, and acidity; endure osmotic stress; and survive mild preservation treatment measures commonly used to control the growth of organisms in food [25].
L. monocytogenes can be found in a wide variety of raw and processed foods including dairy products, raw meat, vegetables, and fishery products, and all these food products have all been associated with Listeria contamination and with outbreaks or sporadic cases of listeriosis [12, 26–29]. For instance, in raw milk and the dairy environment, the source of L. monocytogenes contamination is mainly from poor silage and bedding [30, 31]. L. monocytogenes is transmitted from man to man or from animal to man mostly through ingestion of the organism with contaminated raw milk, dairy products, and raw meat. Hence, these food products have been connected to several outbreaks of listeriosis [32].
Despite the poor surveillance programs and lack of epidemiological data to establish a comprehensive incidence rate of human listeriosis in Africa, there are some studies that reported the occurrence of L. monocytogenes in foods from African countries [33–36]. Moreover, L. monocytogenes strains resistant to one or more antibiotics have been recovered from food, the environment, and sporadic cases of human listeriosis [37–39]. In Ethiopia, the actual epidemiological data about the prevalence as well as antimicrobial susceptibility pattern of L. monocytogenes are lacking both in the veterinary and public health sectors. Moreover, there is no published research work done on the prevalence and antimicrobial susceptibility patterns of L. monocytogenes and its strains in foods of bovine origin, particularly in raw milk and meat in Mekelle City, Ethiopia. Hence, this study was stipulated to assess the prevalence of L. monocytogenes and determine the antimicrobial susceptibility of L. monocytogenes from raw milk and raw meat samples of bovine origin collected from different sources in Mekelle City, Ethiopia.
2. Materials and Methods
2.1. Study Area
Mekelle City is the capital city of Tigray Regional State, which is located at 39° 29′E and 13°−30′N at an average altitude of 2000 m.a.s.l. and 783 km far from Addis Ababa, Ethiopia. The climate of the study area conforms to that of the Ethiopian Highlands. The mean annual rainfall is 619 mm and is bimodal with a short rainy season occurring from March to May and another from middle September to February. The annual minimum and maximum temperature is 11.8°C and 29.9°C, respectively [40]. It is administratively divided into seven subcities and is subdivided into 33 Kebelles. The livestock population of Mekelle city includes 24,419 cattle, 3372 goats, 4935 sheep, 2872 horses, 216 mules, 3080 donkeys, and 293 camels. The total number of dairy farm in Mekelle city is 1515 (comprising of 5634 cross breeds and 13,104 local breeds), and the number of intensive and semi-intensive smallholder dairy farms is 833 [41]. There are two slaughter houses in Mekelle City (Figure 1).
2.2. Study Design and Study Population
A cross-sectional study was conducted from December 2016 to June 2017 in Mekelle City, Ethiopia. The study population comprised of purposively selected milking dairy cows and slaughtered cattle found in Mekelle City. Raw milk and meat samples were collected from randomly selected dairy farms, milk shops, and cafeterias for bovine milk and abattoir, butcher shops, and restaurants for beef.
2.3. Sample Size and Sample Collection
Accordingly, a minimum sample size of 384 was calculated; however, to increase the precision of the estimation, the sample size was doubled to a total of 768 which was distributed to an equal proportion to both raw milk and meat samples. Hence, a total of 768 samples, comprised of milk (n = 384) and meat (n = 384), were collected using a purposive random sampling technique. The nonlactating cows were excluded. Based on the availability of milk sample sources, 30 mL of bovine milk samples was collected from dairy farms (n = 166), shops (n = 120), and cafeterias (n = 98). At the same time, 25 g of bovine meat samples were collected from the abattoir (n = 240), butcher shops (n = 84), and restaurants (n = 60) using sterile plastic bags. All samples were labeled and transported with icebox to the Molecular Biotechnology Laboratory of College of Veterinary Sciences, Mekelle University, at the date of collection and stored at 4°C till further analysis was done.
2.4. Enrichment, Culturing, and Identification
The procedure described by the International Organization for Standardization [43] was used for milk samples for the isolation and identification of L. monocytogenes. While the techniques recommended by the International Standards Organization [44, 45] and the French Association for Standardization [46] with minor modification were employed for meat samples.
The raw milk sample was thoroughly mixed and inoculated into Listeria Enrichment Broth (LEB) at 1:9 ratios. After 48 h of incubation at 30°C, the culture from LEB suspensions was inoculated into Secondary Fraser Enrichment Broth (SFEB) and incubated at 35°C for 24 h. Similarly, for the meat samples, 1:9 proportion of sample to LEB was placed in sterile stomacher plastic bags and homogenized for 1 min in laboratory stomacher (Lab Blender 400, Seward Medical, London, England). Then, the resulting suspension was incubated for 48 h. After 48 h of incubation, 0.1 mL of the culture was transferred to previously prepared SFEB and then incubated for 24 h [47].
After 24 h of incubation, aliquots of positive SFEB medium with black/dark brown or dark green color were taken and streaked onto Listeria Oxford Agar Plates containing selective media with the manufacturer’s supplements, and the plates were incubated at 35°C for 24–48 h. Then, the growth of Listeria species on Listeria Oxford Agar Plate was examined.
Suspected Listeria colonies were subcultured onto Tryptone Soya Agar (TSA) plate and incubated at 37°C for 18–24 h. Those suspected Listeria colonies were characterized by using Gram’s staining, oxidase test, characteristics of hemolysis [47], carbohydrate utilization (xylose and rhamnose), and Christie, Atkins, Munch–Peterson (CAMP) test following standard methods [45, 47]. All culture media and selective supplements were from Oxoid (Basingstoke, UK).
2.5. Genomic DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
The DNA extraction was performed using the phenol-chloroform method for all the isolates from samples of milk and meat according to Sambrook and Russel [48], and the quality and quantity of the extracted DNA were estimated using agarose gel electrophoresis and ultraviolet (UV) spectrophotometer, respectively.
Following preliminary bacteriological identification and genomic DNA extraction of L. monocytogenes isolates, PCR amplification was performed. The PCR condition that was used in this study has been explained previously by Nayak et al. [49], and the sequence of primers mentioned by Bubert et al. [50] for L. monocytogenes and Bobade, Warke, and Kalorey [51] for its serovars wase used. A standard strain of L. monocytogenes ATCC 7644 and nuclease-free ultrapure water were used as positive and negative controls, respectively. The primers used for the PCR reaction were F-3′-TTA TAC GCG ACC GAA GCC AAC-5′ and R-3′-CAA ACT GCT AAC ACA GCT ACT-5′ for amplification of Iap gene of L. monocytogenes (660 bp); F-5′-AGG GCT TCA AGG ACT TAC CC-3′ and R-5′-ACG ATT TCT GCT TGC CAT TC-3′ for amplification of Imo0737 gene of L. monocytogenes serovar 1/2a (691 bp); F-5′-AGC AAA ATG CCA AAA CTC GT-3′ and R-5′-CAT CAC TAA AGC CTC CCA TTG-3′ for amplification of ORF2819 gene of L. monocytogenes serovar 1/2b (471 bp); and F-5′-AGT GGA CAA TTG ATT GGT GAA-3′ and R-5′-CAT CCA TCC CTT ACT TTG GAC-3′ for amplification of ORF2110 gene of L. monocytogenes serovar 4b (597 bp).
Amplification was carried out with thermal cycling conditions of an initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 50°C for 1 min, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min for the detection of L. monocytogenes. But the detection of serovars was optimized with cycling conditions of an initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 15 s, and extension at 72°C for 75 s, with final extension at 72°C for 10 min. Finally, the PCR products along with DNA molecular weight marker (3B Black Bio Biotech) were separated by running on a 2% (w/v) agarose gel containing 0.3 mg/mL ethidium bromide. Electrophoresis was conducted in a horizontal equipment system for 120 min at 90 V using 1X TAE buffer. The amplicons were visualized under a gel documentation system, and their molecular weights were estimated.
2.6. Antimicrobial Susceptibility Testing
All the phenotypically and molecularly characterized isolates of L. monocytogenes were tested for antibiotic susceptibility patterns. The method applied for the in vitro antimicrobial susceptibility testing of L. monocytogenes isolates was the agar plate antibiotic disk diffusion method using Bauer et al. [52] technique. The following 13 antimicrobial disks (which belong to seven classes of antimicrobials) (Himedia Laboratory Pvt., Limited, Mumbai, India) with their concentrations given in parentheses were used in the antibiogram testing: penicillin class antimicrobials (amoxicillin [25 μg], ampicillin [10 μg], cloxacillin [5 μg], methicillin [30 μg], and penicillin G [10 μg]); tetracycline class antimicrobial (tetracycline [30 μg]); fluoroquinolone class antimicrobials (ciprofloxacin [5 μg] and nalidixic acid [30 μg]); lincomycin class antimicrobial (clindamycin [10 μg]); macrolide class antimicrobial (erythromycin [15 μg]); glycopeptide class antimicrobial (vancomycin [30 μg]); and aminoglycoside class antimicrobials (gentamicin [10 μg] and streptomycin [10 μg]). The selection of these antimicrobials was based on the availability and frequent use of these antimicrobials in the study area both in veterinary and human medicine. Standard strains of L. monocytogenes ATCC 7644 and S. aureus ATCC 25923 were used as positive and negative controls, respectively. The results were interpreted as susceptible (S), intermediate (I), and resistant (R) categories based on the critical points recommended by the Clinical and Laboratory Standards Institute [53].
2.7. Data Management and Processing
The data collected from the results of the laboratory investigation were entered into Microsoft Excel Spreadsheet (Microsoft Corp., Redmond, WA, USA) and transferred to STATA version 11.1 for statistical analysis. Descriptive statistics, chi-square test (χ2), and logistic regression were applied to see the associations of the food type and corresponding source as risk factors with that of the occurrence of the positive result; the degree of association was determined using odds ratio (OR) and 95% CI. For all tests, a p-value less than 0.05 was considered statistically significant.
3. Results
3.1. Prevalence of L. monocytogenes
The prevalence of L. monocytogenes, from the total 768 samples, was found to be 3.39%. The sample type prevalence of L. monocytogenes was found to be 4.17% and 2.6% in meat and milk samples, respectively. The prevalence of L. monocytogenes in meat samples collected from abattoir, butcher shops, and restaurants was 1.67%, 8.33%, and 8.33%, respectively. The result was significantly higher (p < 0.05) in butcher shops and restaurants than in the abattoir. The prevalence of L. monocytogenes in the milk samples collected from dairy farms, milk shops, and cafeterias was 1.81%, 2.50%, and 4.08%, respectively. However, there was no statistically significant difference (p > 0.05) in the prevalence of L. monocytogenes among the various sources of milk samples (Table 1).
| Sample type | Place of collection | No. of examined samples | No. of positive samples (%) | 95% CI | p-Value |
|---|---|---|---|---|---|
| Milk | Farms | 166 | 3 (1.81) | 0.0–1.00 | 0.531464 |
| Shops | 120 | 3 (2.50) | 0.27–7.02 | ||
| Cafeterias | 98 | 4 (4.08) | 0.51–10.55 | ||
| Total | 384 | 10 (2.6) | — | ||
| Meat | Abattoir | 240 | 4 (1.67) | 0.27–4.90 | 0.005659 |
| Butchers | 84 | 7 (8.33)a | 1.24–19.62 | ||
| Restaurants | 60 | 5 (8.33) | 0.84–17.87 | ||
| Total | 384 | 16 (4.17) | — | ||
| Grand total | 768 | 26 (3.39) | — | 0.0417 | |
- Abbreviations: CI, confidence interval; %, percent of prevalence.
- aThere was a statistically significant difference (with p-value <0.05).
3.2. PCR Confirmation of L. monocytogenes Isolates and Serotyping
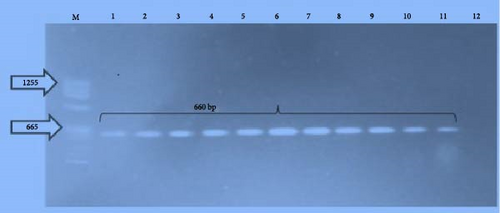
All the phenotypically characterized L. monocytogenes isolates were also molecularly confirmed as L. monocytogenes using species-specific primer, which targets Iap gene as shown in Figure 2.
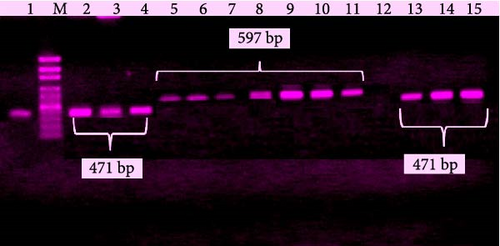
Moreover, the molecularly confirmed L. monocytogenes isolates were further identified at serovar levels using serovar-specific primers with the help of multiplex PCR. The serovars that were identified belonged to 1/2b (38.5%) and 4b (61.5%) as shown in Figure 3.
3.3. Antimicrobial Susceptibility Pattern
The antibiogram profile results indicated as large proportions of the isolates of this study were found to be highly susceptible to ampicillin (88.46%) and vancomycin (84.62%). However, the isolates had shown the highest level of resistance against nalidixic acid (96.15%). The highest intermediate was observed to be amoxicillin (57.69%) (Table 2). Moreover, 34.62% and 42.31% of the isolates developed resistance for two and more than two drugs, respectively, as shown in Figure 4.

| Antimicrobial agents | Interpretations | ||
|---|---|---|---|
| No. of susceptible (%) | No. of intermediate (%) | No. of resistance (%) | |
| Penicillin G | 13 (50.00) | 10 (38.46) | 3 (11.54) |
| Amoxicillin | 9 (34.62) | 15 (57.69) | 2 (7.69) |
| Ampicillin | 23 (88.46) | 3 (11.54) | 0 (0) |
| Vancomycin | 22 (84.62) | 4 (15.38) | 0 (0) |
| Ciprofloxacillin | 20 (76.92) | 6 (23.08) | 0 (0) |
| Streptomycin | 11 (42.31) | 12 (46.15) | 4 (15.38) |
| Nalidixic acid | 0 (0) | 1 (3.85) | 25 (96.15) |
| Gentamycin | 21 (80.77) | 5 (19.23) | 0 (0) |
| Methicillin | 19 (73.07) | 7 (26.92) | 0 (0) |
| Erythromycin | 15 (57.69) | 9 (34.62) | 2 (3.85) |
| Clindamycin | 19 (73.07) | 6 (14.93) | 0 (0) |
| Cloxacillin | 17 (65.38) | 7 (26.92) | 2 (7.69) |
| Tetracycline | 5 (19.23) | 7 (26.92) | 14 (53.85) |
4. Discussion
The overall prevalence of L. monocytogenes from foods of bovine origin was found to be 3.39%, which was characterized and confirmed both phenotypically and molecularly. The current finding is in line with the findings of Kramarenko et al. [54] (2.6%); Reda et al. [55] (2.94%); Ndahi et al. [56] (4%); Walker, Archer, and Banks [25] (4.1%); Morobe et al. [57] (4.3%); Mengesha et al. [58] (4.8%); Molla, Yilma, and Alemayehu [59] (5.1%); Leong, Alvarez-Ordóñez, and Jordan [60] (5.3%); Gebretsadik et al. [61] (5.4%); Garedew et al. [62] (6.25%); and Hawaz, Taye, and Muleta [63] (7.08%). However, it is lower than the findings of Wang et al. [64] (12.4%), Khen et al. [65] (17%), Zafar et al. [66] (26.66%), and Rahimi, Ameri, and Momtaz [67] (32.7%). However, it is higher than the report of Nayak et al. [68] (1.5%).
The meat sample–based prevalence of L. monocytogenes was found to be 4.17%. This is in agreement with the findings of Gebretsadik et al. [61] (2.6%), Nastasijevic et al. [69] (3.3%), Ndahi et al. [56] (3.92%), and Garedew et al. [62] (6.66%). However, it is lower than the reports of Reda et al. [55] (8%), Shen et al. [70] (9.6%), Wang et al. [64] (10.3%), Teixeira et al. [71] (12%), Khen et al. [65] (14%), Awadallah and Suelam [72] (15%), Indrawattana et al. [73] (15.4%), Kramarenko et al. [54] (18.7%), and Demaîtrea et al. [74] (45.6%). However, it is higher than the finding of Nayak et al. [68] (0%).
Likewise, the milk sample–based prevalence of L. monocytogenes was found to be 2.6%. This report is in agreement with the reports of Rawool et al. [75] (0.41%); Rahimi, Ameri, and Momtaz [67] (1.1%); Garedew et al. [62] (4.0%); Muhammed et al. [76] (4%); Nayak et al. [68] (4.0%); Hamdi et al. [77] (4.29%); Kevenk and Gulel [78] (5%); and Kalorey et al. [79] (5.1%). However, it is lower than the reports of Seyoum et al. [80] (5.6%); Obaidat and Stringer [81] (7.54%); Gebretsadik et al. [61] (13%); Morobe et al. [57] (5.3%); Reda et al. [55] (8%); James et al. [82] (8.8%); Kramarenko et al. [54] (18.1%); Jamali, Radmehr, and Thong [83] (21.7%); and Zafar et al. [66] (40%). In general, the variation both in the overall and sample-wise prevalence rates of L. monocytogenes might be due to differences in the sample types of foods of bovine origin, sources of the samples, processing plants, approaches of sample collection, sample size, methodological approaches, isolation and identification techniques, prevalence calculation/interpretation, geographical locations, hygienic conditions, handling and transportation of samples, and contamination rates from utensils and personnel.
The serovars that were identified in the current study belonged to 1/2b and 4b. Kevenk and Terzi Gulel [78] and Rawool et al. [75] also reported these serovars in addition to some others. In general, the most common causes of human listeriosis among the 13 serotypes of L. monocytogenes are 1/2a, 1/2b, and 4b, and of these, serotype 4b has been related to the most recent outbreaks of listeriosis, and serotypes 1/2a and 4b are commonly reported in animals [14–20, 32]. The presence of molecular serogroup 1/2b and 4b isolates (potential serotype 4b) in food of bovine origin may pose a great health threat since L. monocytogenes 4b has caused numerous human listeriosis outbreaks [84].
In the present study, the antimicrobial susceptibility results indicated as large proportions of the isolates were found to be highly susceptible to ampicillin (88.46%) and vancomycin (84.62%). However, the isolates had shown the highest level of resistance against nalidixic acid (96.15%). The highest intermediate was observed in amoxicillin (57.69%). This is in agreement with the reports of Hawaz, Taye, and Muleta [63] who reported the highest level of resistance against nalidixic acid (100%), but 100% of the L. monocytogenes isolates were sensitive to vancomycin; Garedew et al. [62] who reported a high degree of resistance against nalidixic acid (50%) and tetracycline (37.5%) but the highest level of sensitivity to vancomycin (100%); Khen et al. [65] who reported 97% of susceptibility to ampicillin and vancomycin; Shen et al. [70] who reported 100% of susceptibility to vancomycin; Zafar et al. [66] who reported 100% of susceptibility to ampicillin and vancomycin; James et al. [82] who reported 100% of susceptibility to ampicillin, gentamycin, and vancomycin and 24.2% of resistance against tetracycline; Kevenk and Gulel [78] who reported a high level of sensitivity to ampicillin (78.9%), penicillin G (77%), erythromycin (71.2%), and vancomycin (67.3%); and Rahimi, Ameri, and Momtaz [67] who reported 100% of susceptibility to vancomycin and 96.4% of resistant against nalidixic acid. This is contradicted by the findings of Welekidan et al. [85] who reported a high degree of resistance against amoxicillin (50%) and vancomycin (50%); Jamali, Radmehr, and Thong [83] who reported a high degree of resistance against clindamycin (100%), ampicillin (95.7%), erythromycin (95.7%), penicillin (91.3%), tetracyclin (82.6%), streptomycin (78.3%), and vancomycin (69.7%); and Garedew et al. [62] who reported a high degree of resistance against penicillin (66.7%) and the highest level of sensitivity to amoxicillin (100%), cloxacillin (100%), and gentamicin (100%). The current report is similar to the findings of Wang et al. [64] who reported 13.6% of intermediate response to ciprofloxacin. Moreover, 23.10%, 34.62%, and 42.31% of the isolates showed resistance to one, two, and more than two drugs, respectively. This finding is lower than the findings of Obaidat and Stringer [81] who reported 96.9% of multidrug resistance. However, it is higher than the report of Garedew et al. [62] (16.7%) and Escolar et al. [86] who reported a 2% of resistance to two antibiotics and 2% resistance to three antibiotics and Kevenk and Terzi Gulel [78] who reported that 15.3% of the isolates were resistant to only one drug and 36.5% were resistant to multiple drugs. However, it is lower than the reports of Wang et al. [64] who reported 72.3% of multiple drug resistance. In general, L. monocytogenes is usually susceptible to a wide range of antimicrobials. Nevertheless, the evolution of bacterial resistance toward antibiotics has been accelerated considerably by the selective pressure exerted by the overprescription of drugs in clinical settings and their heavy use as promoters in animals husbandry [39]. Moreover, the genes responsible for antibiotic resistance could be transferred through movable genetic elements such as conjugative transposons, mobilizable plasmids, and self-transferable plasmids to other foodborne bacteria in the gastrointestinal tract. In Listeria spp., efflux pumps have also been reported as the resistant mechanism [87]. Hence, the use of antimicrobials in veterinary medicine is the main cause of the development of AMR foodborne bacterial pathogens including L. monocytogenes, as AMR pathogens can easily be transported from animal to human via food consumption [88].
5. Limitations
Even though the present study is among the valuable studies to offer valuable information on the prevalence and antimicrobial resistance pattern of L. monocytogenes, it is not without limitations. The limitations of the present study were as follows: it did not address the knowledge, attitude, and practice of the farm owners, workers, and consumers towards foodborne pathogens and antimicrobial usage practices, contamination rates from utensils and personnel, and seasonal variations and did not represent the nation-wise prevalence rate and antimicrobial resistance profile of L. monocytogenes. Moreover, it did not include a minimum inhibitory concentration analysis, and the isolates were not well characterized molecularly both for the organism and its serotype and drug-resistant genes such as hly gene, sequencing of the 16S rDNA or iap genes, prs, lmo1118, phage typing, DNA restriction enzyme, nucleic acid sequencing-based typing, and microarray analysis.
6. Conclusions
In conclusion, the current study revealed the occurrence of L. monocytogenes with different pathogenic serovars (1/2b and 4b) in raw meat and milk samples of bovine origin and the development of drug resistance in Mekelle City. Hence, it has an important public health implication when antimicrobial-resistant L. monocytogenes is present in raw foods, especially in developing countries like Ethiopia where there is a very common practice of consumption of raw meat and milk products and widespread and uncontrolled use of antimicrobials. Thus, this indicates the need to practice adequate cooking of bovine origin foods before consumption and rational use of antimicrobials both in veterinary and human treatment regimens with regular surveillance of antimicrobial resistance to combat multidrug resistance.
Ethics Statement
This study was reviewed and approved by the Research Ethics Committee of the College of Veterinary Sciences, Mekelle University. Owners, managers, and workers of the different sites were informed of the procedures and significance of the study. Each data and analysis results were kept confidential and communicated to concerned bodies. Any participants who were not volunteers were not forced to be included.
Disclosure
A preprint has previously been published with doi.org/10.1101/2021.11.16.468824 at bioRxiv, the preprint server for biology [89]. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
All the authors, T.H., G.G., Y.T., M.A., and N.A., contributed to conceptualization, design, funding acquisition, carrying out the study, data curation, analyzing and interpreting the results, and writing and revising the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Norwegian Agency for Development Cooperation (NORAD) and One Health Central and Eastern Africa (OHCEA) project funds of College of Veterinary Sciences, Mekelle University.
Acknowledgments
The authors would like to acknowledge the owners, managers, and workers of the different dairy farms, abattoir, butcher shops, restaurants, milk whole sellers, and cafeterias of the study site for their keen interest and cooperation during the collection of the samples and to the College of Veterinary Sciences staff members who directly or indirectly helped us during the research period.
Open Research
Data Availability Statement
All the data generated or analyzed during this study are included within the article.




