Molecular Systematics and Divergence of Tor and Neolissochilus Fishes (Cypriniforms: Cyprinidae) from Southeast Asia and South China
Abstract
In Southeast Asia, mahseer fishes, such as the species of Tor and Neolissochilus, are significant native commercial fish. Their phylogeny and categorization have a convoluted history. In this study, the molecular systematics and divergence of Tor and Neolissochilus fishes from Southeast Asia and South China were examined using the partial or complete sequences of four mitochondrial genes (cytochrome oxidase I, cytochrome b, 16S, and ND4). This study substantiated the monophyly of Tor and supported N. benasi as an independent genus. The result supported T. laterivittatus as a synonym of T. sinensis and T. dongnaiensis as a synonym of T. tambra. In addition, we presented the high diversity and species crypticity of these two taxa in Southeast Asia. The divergent time estimation indicated Tor and Neolissochilus species originated in the early Miocene (about 16.73 Ma), and the divergence of the genus Tor and other species of the genus Neolissochilus began at about 12.86 Ma.
1. Introduction
The cyprinid fishes belonging to the genera Tor and Neolissochilus are commonly known as mahseer, which have been classified under the subfamily Torinae of Cyprinidae after extensive discussion on the phylogenetics of Cypriniformes [1, 2, 3, 4]. They are widely distributed in Pakistan, India, Bhutan, Bangladesh, Nepal, Sri Lanka, Myanmar, Laos, Vietnam, Thailand, Cambodia, and other countries in South Asia and Southeast Asia. In China, it is only distributed in Yunnan and Tibet. They live in a bunch of drainages, from the Yarlung Tsangpo River, Irrawaddy River, Salween River, and Mekong River to the Red River, as well as drainages in the Malay Peninsula, Sumatra, Java, Borneo, etc. They are omnivorous fish in the middle and lower layers. The adult mostly resides in the calm areas of the main river, such as backwater bays, large tributaries, or deep-water regions with sluggish currents. On the other hand, subadults or juveniles are generally found in the higher-altitude water bodies of the tributaries. They exhibit anadromous movement as part of their reproductive behavior. The majority of these species have significant economic value within their respective geographical regions.
Currently, 17 species are valid in the genus Tor Gray and 32 species in the genus Neolissochilus Rainboth [2]. Phylogenetic relationships, morphological classification, and geographical distribution are closely related between the two fish genera. The main differences in morphology of the two genera are scale size, number of lateral-line scales, number of gill rakers, postlabial groove, presence or absence of a median lobe and its length, which is enlarged to form a median lobe and with a continuous postlabial groove in Tor species, while with no median lobe and interrupted postlabial groove in Neolissochilus species. The species classification of these two genera is still confusing because the shape, size, and length of the median lobe are often highly variable [5], which is related to habitat and the ability to adhere in rapids [6, 7]. Subsequently, tropic polymorphisms were reported in the Neolissochilus species [8, 9]. Laskar et al. [10] emphasized the importance of the number of gill rakers on the first arm of the gill arch in distinguishing the two genera. The two genera have some overlapping morphological and meristic characters and share almost the same distribution area. Simultaneously, the description and identification of the two genera may have greater uncertainty due to the phenotypic plasticity and intraspecies variation affected by different geographical and environmental conditions, as well as the different selection and description of characters by different researchers. Because of these, there has not yet reached a consensus on the classification of species among taxonomists.
Most recent molecular phylogenetic studies on mahseer only used DNA barcoding to look at the variety of species and the validity of Tor and Neolissochilus. Hoang et al. [8] described two new species, T. mekongensis and T. dongnaiensis, based on the cytochrome oxidase I (COI) gene with a genetic distance of 0.7%. Subsequently, a genetic analysis by Walton et al. [11] showed that T. dongnaiensis could not be significantly distinguished from T. tambra in peninsular Malaysia and western Java. While Pinder et al. [12] supported the validity of T. dongnaiensis in their review of global Tor fish, but they considered that T. mekongensis may be a synonym of T. dongnaiensis. Hoang et al. [8] also discovered a new species record in South Vietnam, N. stracheyi, and showed that it was clustered with specimens from Laos; and considering that characters such as body color and median lobe have many variations, it is believed that the individuals identified by previous studies [13, 14] could correspond to the Tor-like morph of N. stracheyi. Lalramliana et al. [15] used DNA barcoding to describe a new species, N. kaladanensis, distributed in northeast India, and discovered that the conclusions supported by the morphological and molecular data of the relationship between N. baoshanensis and N. stracheyi were inconsistent; reasons for this inconsistency may include the fact that the species itself is synonymous, species identification errors, or limitations of DNA barcoding. Nguyen et al. [16] explored the phylogeny, evolution, and biogeography of six species of Tor and five species of Neolissochilus using three mitochondrial genes and showed that these two genera are sister groups of each other. Subsequent studies found that T. hemispinus and T. qiaojiensis were embedded with a clade containing Neolissochilus species, which did not support the monophyly of these two genera [15, 17]. It should be noted that both the molecular markers and the number of species involved in these studies, while the geographical coverage is small and phylogenetic relationships may not be very accurately inferred, are likely to change significantly as the species coverage or the strength of the molecular markers increases.
Currently, few studies focus on the biogeography of Tor and Neolissochilus. Only Nguyen et al. [16] proposed two hypotheses on the present distribution of mahseers: vicariance fragmentation and range expansion and speculated that the division between Neolissochilus and Tor probably occurred 7.6–3.8 Ma, or during the Miocene epoch. Most studies focused on higher-scale taxa and did not discuss these fishes. For example, Yang et al. [4] reconstructed the phylogenetic relationship of the Cyprininae sensu lato based on the RAG1 gene and mitochondrial genes and subsequently deduced that the tetraploidization and hexaploidization events of Torini likely occurred before the diversification (32.5 Ma) of tetraploid Torini and after the origin (18.1 Ma) of the now hexaploid Torini, respectively [18]. Phylogenetic trees based on different datasets revealed different topologies for the relationships between Tor and Neolissochilus. Therefore, it is necessary to construct a stable phylogenetic tree on a small scale and focus on the phylogenetic relationships and evolution of the study taxa.
The species belonging to the genera Tor and Neolissochilus have a broad distribution in Southeast Asia and South Asia. This research specifically focused on Southeast Asia and the Yunnan Province of South China, with a comprehensive examination of the species belonging to the Tor and Neolissochilus. By performing a molecular systematic study and divergence time estimation, we aim to inspect the validity of the species, understand their phylogenetic relationships, and explore their origin and evolution.
2. Materials and Methods
2.1. Sample Collection
Sampling and fieldwork in Yunnan province and Southeast Asia were carried out from 2005 to January 2023 by the Kunming Institute of Zoology, CAS. All specimens were collected as part of routine surveys. All fish handling and experiments complied with the relevant laws of the Chinese Laboratory of Animal Welfare and Ethics (GB/T 35892-2018). All fished individuals were rapidly euthanized by 10–100 mg/L anesthetic clove oil. The right pelvic fin was severed and preserved with pure alcohol and stored in the refrigerator at −80°C. Specimens were preserved by immersion in 75% alcohol or 10% formalin. General measurements and counts refer to Rainboth [19] and Laskar et al. [10]. A total of 153 individuals identified as Tor and Neolissochilus in the wild were selected for PCR; the genus Tor includes 41 specimens (5 valid plus 3 undescribed), and Neolissochilus includes 112 specimens (10 valid plus 4 undescribed). A further 53 sequences of the genera Tor and Neolissochilus were downloaded from the NCBI GenBank database and used as the ingroup together with the sequences obtained in this study. The ingroup taxa finally included 13 valid species and 3 undescribed species of genus Tor, 17 valid species, and 4 undescribed species of Neolissochilus. We retrieved reference sequences of 45 species from the NCBI GenBank database to be used as an outgroup in this study. Sample and sequence information used in the current study can be seen in Supplementary table 1.
2.2. DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted from the fin clips that were preserved in pure ethanol. COI, cytochrome b (Cyt b), 16S ribosomal RNA (16S), and NADH dehydrogenase subunits 4 (ND4) were used in this study. DNA was extracted using the DNA Isolation Mini Kit (Vazyme Biotech Co., Ltd.). PCR was carried out in a total volume of 25 μL, which contained 1 μL (10 μM) of each Primer, 1 μL Template DNA, 9.5 μL ddH2O, and 12.5 μL 2 × Taq Master Mix (Vazyme Biotech Co., Ltd.). Primers used for sequencing are provided in Supplementary table 2. Two pair primers of 16S were used to amplify and assemble the full-length gene sequence, and primers for the ND4 gene were the universal primers designed for this study by Sangon Biotech (Shanghai) Co., Ltd. PCR conditions for COI and Cyt b fragment were: initial denaturing at 92°C for 3 min, followed by 35 cycles of denaturing at 94°C for 45 s, annealing at 52°C for 45 s, and extension at 72°C for 60 s, and a final extension at 72°C for 5 min, then hold at 10°C; for 16 S: initial denaturing at 95°C for 3 min, followed by 30 cycles of 94°C for 30 s, 58–62°C for 30 s, and 72°C for 60–90 s, and a final extension at 72°C for 5 min, then hold at 10°C; for ND4 encompassed initial denaturing at 95°C for 5 min, 30 cycles of 94°C for 45 s, 55°C for 45 s, extension at 72°C for 90 s, then followed by a 10 min final extension at 72°C. Purification and sequencing in both forward and reverse directions were undertaken at Sangon Biotech (Shanghai) Co., Ltd.
2.3. Sequence Alignment and Molecular Systematics
Sequences were manually proofread and assembled using SeqMan in the DNASTAR Lasergene v7.1 (DNASTAR, Inc., USA) and aligned by MUSCLE in MEGA7 [20]. To evaluate the phylogenetic performance of each gene and concatenated sequences, we used Lamp in the IQ-TREEv2.2.0 [21] to perform Likelihood Mapping analysis [22], and the Incongruence Length Difference Test was used to test the homogeneity in PAUP ∗4.0 [23]. Sequence datasets were analyzed in DnaSP v6 [24] to get the sequence characteristic information. The genetic distance between the species groups was calculated by averaging pair-wise comparisons of sequence across all individuals as implemented using MEGA 7.0.
Three methods were used to construct the phylogenetic tree for the concatenated dataset of four genes: maximum likelihood (ML), Bayesian inference (BI), and maximum clade credibility tree (MCC). All operations of ML and BI were done in PhyloSuite v1.2.2 [25]. Node support was determined by setting 1,000 bootstrap (BS) replicates in ML and 20,000,000 generations of Markov chains, sampling every 2,000 generations to yield 10,000 trees in BI. MCC analysis was performed in BEAST V1.10.4 [26]. The best partition scheme and evolutionary models were selected using PartitionFinder2 [27] with all algorithms and AICc criteria.
2.4. Divergence Time Estimation
Divergence time analysis used Opsariichthys bidens and six species of genus Labeo as outgroups in this study, and the tree topology yielded by ML analysis was used for inferring the divergence time. Two fossil calibrating points were selected to calibrate the divergence time. One was the separation of subfamilies Cyprininae and Leuciscinae in the mid-Oligocene (27.7 Ma) [28]; one was the oldest African Labeo-like fossil from Loperot, Kenya, dating to the early Miocene (17.0 Ma). The filtered sequence dataset was parameterized using BEAUTi in the BEAST package to generate the input file. The analysis was performed using Unlink Subst. Models are carried out under a relaxed clock lognormal model. Yule model was selected as a prior. The Markov chain was run for 20,000,000 generations and sampled every 2,000 generations, then the first 10% of the trees were discarded as burn-in. The log results were examined in Tracer 1.7.2 [29]. The maximum credibility tree was summarized using TreeAnnotator in BEAST and visualized in FigTree 1.4.4 and iTOL (https://itol.embl.de/).
3. Results
3.1. Gene Characteristics and Variation
The first dataset was assembled for phylogeny, including all the sequences obtained in this study, 251 sequences in total (45 outgroup and 206 ingroup sequences). The aligned length of the first concatenated dataset (COI + Cyt b + ND4 + 16S) was 4,196 bp, with 1657 parsimony-informative sites, 308 singleton sites, and 2,231 constant sites. The average base composition was 26.6% T, 23.0% C, 29.5% A, and 20.9% G, and the estimated Ti/Tv was 4.0. The second dataset included the concatenated sequences of O. bidens, genus Labeo fishes, and selected representatives from the ingroup for divergence time analysis (Supplementary table 1). It included 53 sequences, and the aligned sequence length was 4,224 bp, with 1,000 parsimony-informative sites, 382 singleton sites, and 2,842 constant sites. The average base composition was 26.4% T, 23.1% C, 29.4% A, and 21.0% G, and the estimated Ti/Tv was 4.6.
3.2. Phylogenetic Relationships
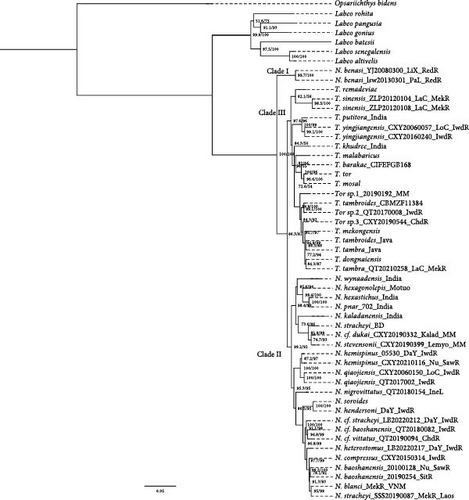
The topologies of the reconstructed phylogenetic trees in ML and BI were similar. The result illustrated the species clustering situation and the genera Tor and Neolissochilus formed a paraphyletic group located at the top of the phylogenetic tree, which, together with Labeobarbus and Varicorhinus in Africa, constituted a clade with high support (Figure 1). The samples of Tor and Neolissochilus have been divided into three strongly supported evolutionary clades (BS = 100). The Clade Ⅰ (red branch) located at the base of both genera was composed only of Neolissochilus benasi, but all other species in Neolissochilus were clustered into Clade Ⅱ (blue branch), which is the sister group to Clade Ⅲ (green branch) that contained all the samples from Tor. All phylogenetic analyses corroborated Tor as a monophyletic group with no obvious geographical structure; some species, including T. malabaricus, T. khudree, and T. barakae confined to India were not well elucidated, and they appeared to be multibranched in the BI tree (not shown in this article). Meanwhile, the phylogenetic position of T. remadeviae may be changed due to outgroup selection. Among the two species in the Dongnai River, T. mekongensis had a clear phylogenetic branch, while T. dongnaiensis and a part of T. tambra clustered together. Furthermore, T. tambra and T. tambroides of Sunda Island are obviously mixed in the phylogenetic tree, excluding the Mekong basin. T. putitora from India and T. yingjiangensis from the Irrawaddy River basin were the most closely related, and both had clear phylogenetic status (BS = 100). Phylogenetic reconstructions did not support the monophyly of Neolissochilus. It was obviously divided into two clades (BS = 100), one of which was Clade Ⅱ and contained the type species of Neolissochilus. Clade Ⅱ can be further divided into two subclades, one containing species closer to South Asia, i.e., India, Motuo in Tibet, and the region bordering India in Myanmar, located at the base of the phylogenetic tree of this genus; the other contained only Southeast Asian species, ranging from Yunnan in China to the island countries of Southeast Asia. The species identified as N. stracheyi downloaded from NCBI were scattered across the branches of this genus, with individuals from the Mekong Basin at the top and having the closest evolutionary relationship to N. baoshanensis. It was notable that an individual named N. blanci was mixed in, but it was actually more likely to be N. stracheyi. Neolissochilus hemispinus contained the Nujiang River, Longchuanjiang River, and Dayingjiang River individuals, which were resolved to be most closely related to N. qiaojiensis. The phylogenetic tree derived from the second dataset (Figure 2) displayed interspecific relationships more concisely and accurately.

3.3. Divergence Time Estimates
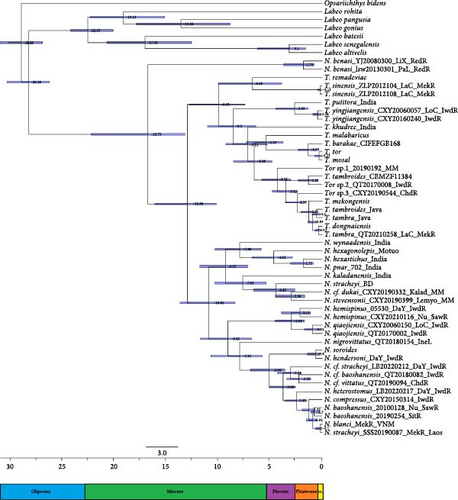
Divergence time estimates for major nodes are provided in Figure 3. This analysis revealed that the 95% highest posterior density (HPD) interval is 13.25–20.61 Ma, indicating that the common ancestor of Tor and Neolissochilus most likely diverged approximately 16.73 Ma of the mid-Miocene (Supplementary figure 1). N. benasi had a separate evolutionary pathway, while Tor and other Neolissochilus species began to differentiate around 12.86 Ma. The common ancestor of Tor likely diverged around 9.92 Ma. The main speciation event occurred in the Pliocene–Pleistocene.
4. Discussion
4.1. Taxonomic Classifications
Based on molecular data, most species’ status was highly consistent with previous studies. The results presented herein encompassed 13 nominal species of Tor with three undescribed species, as well as 17 nominal species of Neolissochilus with four undescribed species. Due to the inadequate genetic data and morphological descriptions, the undescribed species are labeled as Tor sp.1, Tor sp.2, Tor sp.3, Neolissochilus cf. dukai, Neolissochilus cf. stracheyi, Neolissochilus cf. vittatus, and Neolissochilus cf. baoshanensis in the present study. Given taxonomic ambiguities, some species validity must be cautiously interpreted.
Within the genus Tor, a new species, T. laterivittatus, was described from the Nanla River, a tributary of the Lancang River, and a dark longitudinal stripe along the side of body being used as a key character to distinguish it from T. sinensis [14]. Subsequently, Roberts [13] examined and described the adults and subadults of T. sinensis with a deep, dark mid-lateral stripe from the Mekong basin in Laos and placed T. laterivittatus as a synonym of T. sinensis. In the present study, topotypic material (Figure 4(a)) from the Menglun, Nanla Rive, matched the original description of T. laterivittatus well and was genetically identical to that of T. sinensis from the Lancang River in Yunnan and the Mekong River in Laos (Figure 4(b)), six of which were collected in the same small area as the type locality. Therefore, our result supported the assertion of Roberts [13] that T. laterivittatus is a synonym of T. sinensis.


The taxonomic identification between T. tambra and T. tambroides has been confusing. Current knowledge indicates the main difference between them is the length and shape of the median lobe; one with a short square median lobe was identified as T. tambra, while T. tambroides had a longer median lobe [30]. The addition of two more newly described species, T. dongnaiensis and T. mekongensis, has intensified the taxonomic discussion. Less conspicuous morphological characteristics, such as the length of the median lobe, set these two species apart from T. tambra and T. tambrodies [8]. In our phylogenetic tree (Figure 1), T. dongnaiensis was nested in T. tambra from the Mekong basin, and phylogeny does not support its validity. The result was consistent with Walton et al. [11], who revealed a mixture of long-lobe and short-lobe morphotypes of the “T. tambroides” and considered the validity of T. mekongensis and T. dongnaiensis require further confirmation. We supported T. dongnaiensis as a synonym of T. tambra, but the monophyly and validity of T. mekongensis were obvious. The phylogenetic tree also showed that T. tambra had distinct geographic differentiation between the Mekong basin from China to Vietnam and the Sunda Islands. A similar result was mentioned by Nguyen et al. [16], which showed T. douronensis ( = T. tambra) formed three distinct lineages, these being the Mekong river system from China to Vietnam, Sumatra Island in Indonesia, and Sarawak and Sabah (Borneo Island) in East Malaysia. However, species identification and geographic distribution of T. tambra are still confusing. What is noteworthy is the classification of T. tambra and T. tambroides in the Sunda Islands. Roberts [13] considered that the length and size of the median lobe are highly variable at sexual maturity and treated T. tambroides as junior synonyms of T. tambra in the Malay Peninsula, Sumatra, Borneo, and Java. Nguyen et al. [16] showed that samples from the Pahang River have a longer median lobe and that T. tambroides is a valid species. Recent studies based on mitochondrial genomes showed that species sequences are mixed with topologies similar to this study [31, 32]. Our morphological study observed that individuals of T. tambra in the Mekong basin had a short median lobe, except for T. dongnaiensis ( = T. tambra), which has a long median lobe. Additionally, the samples from Java and Malaysia came together to form a single clade that included T. tambroides. The explanation may require careful consideration of the possibility of identical species, as well as mitochondrial genetic confusion and morphological neutralization caused by hybridization due to the proliferation of aquaculture and trade in Southeast Asia. Although the location of T. tambroides CBMZF11758 was unknown, this study stated that it’s grouped with Tor sp. 2 from the Irrawaddy River. Genetic divergence separates this clade from other T. tambroides and T. tambra species. Lim et al. [31] postulated that either this fish has been misidentified as T. tambroides or is possibly a cryptic species. We consider it to be a cryptic species, similar to the Tor sp. 3. The finding of cryptic species implies that there is likely more complexity to the species diversity of Tor in Southeast Asia.
Some studies have suggested that the species diversity within Neolissochilus may be severely underestimated with multiple undescribed species [33]. Such as the discovery of five new species confined to the Western Ghats [34]. Our analysis also showed that there are more clades in Southeast Asia that have not been clearly identified, including Neolissochilus cf. dukai, Neolissochilus cf. stracheyi, Neolissochilus cf. vittatus, and Neolissochilus cf. baoshanensis, which required more detailed morphological analysis and molecular phylogeny to identify species, due to the roughly primitive description of species and the lack of sequence information in the NCBI database. At the same time, Neolissochilus species are reported to have phenotypic polymorphism and intraspecific variation. Trophic polymorphism was reported in the N. soroides from the Malay Peninsula, N. hendersoni from Penang Island, and N. stracheyi from the Mekong River, mainly manifested in the degree of lip or median lobe development [8, 9]. Obvious intraspecific variations can also be observed about head shape and median lobe size in N. qiaojiensis for the first time in this study. According to the first description, the median lobe of N. qiaojiensis was underdeveloped, being just slightly broader than the sides of the lower lip or forming a transversely membranous lobe [35]. Nevertheless, our study indicates that within this group, there are individuals who possess a surprisingly long median lobe (Figure 5(b)), as well as those who closely resemble the original description (Figure 5(a)). Another species with intraspecific differentiation is N. hemispinus, which was only distributed in the Nujiang River from Yunnan before. However, we identified some individuals collected from the Dayingjiang River and Longchuanjiang River of the Irrawaddy drainage based on the present molecular systematic results (Figure 1). The morphological differences between Nujiang drainage and Irrawaddy drainage were mainly manifested in body shape, the number of scales and fins, etc., such as the number of lateral-line scales (33–35 vs. 27–28), predorsal scales (11–12 vs. 9), the degree of ossification of the last unbranched dorsal-fin rays (less than 1/2 vs. more than 2/3) and transverse scales (27 vs. 20). It is a newly recorded fish in the Dayingjiang River and Longchuanjiang River, which could easily be misidentified as N. qiaojiensis from the same drainage basin. The difference in morphology may be due to natural selection pressures such as habitats and food resources in different water systems, which have led to divergent evolution of species morphological characteristics, but it has not reached the species level. After increasing the sample size of N. hemispinus in the Nujiang River, the morphological differences may be reduced.


Among the genus Neolissochilus, N. stracheyi, as the type species, initially appeared in Mawlamine and Sittwe, Myanmar, at the mouth of the Salween and Kaladan rivers, respectively [36, 37]. Later literature recorded that it was widely distributed throughout major river systems in Southeast Asia, such as the Mekong basins that include Laos, Thailand, Cambodia, and Vietnam; the Chao Phyara and Salween basin; the Tennasserim basin to the Mekong River, the Chindwin River in western Myanmar; and the Nanla River in Yunnan, China [19, 38, 39, 40, 41, 42]. Rahman et al. [43] have used DNA barcoding to identify N. stracheyi (GenBank No. MK572365-67) from Chittagong, Bangladesh, without further explanation. However, this sample, N. stracheyi Bangla, was clustered with Neolissochilus species but far away from N. stracheyi in Laos based on our phylogenetic tree (Figure 1). Since no distribution of Neolissochilus species has ever been documented in Bangladesh, we assumed that the population there represented a distinct species that has not yet been described. Another individual used in Barman et al. [44], N. strachyi India, was grouped with samples from Myanmar and identified as Neolissochilus cf. vittatus, but no matched location information was given. We considered it to be a misidentification of N. stracheyi and may collect from the Chindwin River basin in Manipur, India, which was consistent with the distribution of Neolissochilus cf. vittatus in Myanmar. The three nominal Neolissochilus species, N. compressus, N. stracheyi, and N. baoshanensis, needed to be discussed with caution because of their very low genetic differences from each other. Previous studies also reported that evidence of genetic distance differences based on COI does not support morphological results about some Neolissochilus species, like N. stracheyi and N. baoshanensis [15, 45]. Our research implied that these three species are very closely related and may be the same species. Populations in different drainages may represent different lineages and diverged from each other, but the degree of differentiation has not reached the level of independent species. Further analysis of the geographical distribution, N. compressus was a new species described based on one specimen by Day [46], whose type locality was corrected to the upper Irrawaddy River in Burma, rather than the erroneous Kashmir [47]. Wei et al. [48] published the mitochondrial genome of N. stracheyi (GenBank No. OM203155), which was collected from the Dayingjiang River in Yunnan, whereas according to our phylogenetic tree, this individual is more closely related to N. baoshanensis from the Nujiang River and Sittang River, whose type locality was the Longchuanjiang River and the Nujiang River [49]. The geographical distribution of these three species can be considered to be sympatric, which supported our hypothesis to some extent. Furthermore, it also reminded us to be cautious about the taxonomic status of these N. stracheyi, as the existing phylogenetic inferences were almost based on individuals from the Mekong River, not from the type locality or even type watersheds.
It is well known that molecular-based species delimitation might present challenges in establishing distinct clades for divergence events that occurred recently, as evolving lineages may not have accumulated sufficient mutational differences. [50, 51]. Meanwhile, some studies have shown that the species of Torinae have a hybrid origin [52, 53]. It is difficult to distinguish allopolyploidy from homologous polyploidy based on tree topology when polyploidy occurs in two very close parents [4, 18]. Due to the maternal inheritance of mitochondria, different species may have similar or identical maternal parents, resulting in small genetic differences in mitochondrial genes between species. Hybridization plays an important role in the distribution pattern of genetic diversity, taxon differentiation, and speciation [54]. Among fish, evidence of hybridization is more prevalent in the fresh waters, especially between carps and minnows (Cypriniformes), owing to their high diversity, sympatry, and breeding behaviors [55, 56]. Studies such as high-throughput sequencing and population genetic analysis can further detect hybridization and explain these issues [57, 58].
4.2. Monophyly of Tor and Neolissochilus
The present study represents the most comprehensive phylogenetic study of the genus Tor and Neolissochilus in terms of sample size and molecular information to date. Since the genus Neolissochilus was split off from fish like Tor and Barbodes, it has been thought to be closely related to Tor [19, 59, 60], but later studies have called their phylogenetic positions into question [15, 16, 17]. The topology of the phylogenetic tree in this study is consistent with the results of Zheng et al. [17], who proposed that the N. benasi in Red River might represent a new genus. Tor is a monophyletic group, whereas the monophyly of Neolissochilus is not supported because N. benasi forms a separate clade. The divergence time results further proved that it has an independent evolutionary pathway earlier than other species. It should be emphasized that the validity of its status needs to be supported by detailed morphological work.
4.3. Pattern of Distribution Implications
All currently known species of Tor and Neolissochilus are endemic to the tropical and subtropical areas between the trans-Himalayan region in the northwest and the islands of Southeast Asia. Our calibrated divergence time analysis estimated that these two genera of fish may have originated at 16.73 Ma, well matched with the rapid uplift of the Himalayas (23–19 Ma) [61, 62], and species mainly began to differentiate in the Pleistocene, then speciation in the late Miocene. N. benasi, restricted to the Red River basin, was isolated alone at 16.73 Ma, which coincides with the timing of the formation of the Red River zone, which commenced at 27 Ma and ended at 21–17 Ma [63, 64, 65]. It has been hypothesized that the Red River zone at this stage acted as a potential geographic barrier for species dispersal; any change in the runoff regime could have caused a stronger isolation effect by the Red River at that time [66]. Two major events have been suggested as driving the formation and distribution of various species in Asia since the late Miocene [67]. The first is the oscillation of climate and glacial and interglacial periods that occurred regularly in the Pliocene and Pleistocene. Since the middle Miocene, repeated changes in sea level due to climate change and glaciation have led to significant speciation and faunal succession in Southeast Asia [68, 69]. The second is tectonic activity, including rifting, uplifts of plateaus, and earthquakes. For example, studies have shown that the uplift of the Qinghai—Tibet Plateau and Himalayas may have important effects on the formation, divergence, and distribution of species around the Himalayan area [67, 70]. Meanwhile, these activities also affected the hydrological changes in Southeast Asia. River rearrangement-capture is considered to be an important factor for the isolation, diversification, and cross-basin distribution of fish [71, 72]. The species distribution in the present study was similarly consistent with the above assumptions, and the results supported that land connectivity due to climate fluctuations since the Middle Pleistocene led to the widespread distribution of T. tambra. Moreover, the new distribution of N. baoshanensis in the Sittang River may further hint at runoff changes in the Irrawaddy River, where the two were interconnected in the ancient drainage system. The isolation and divergence brought about by frequent geological and climatic changes in Southeast Asia may be one of the reasons to explain such a rich genetic diversity of the fish of the genera Tor and Neolissochilus.
5. Conclusions
This study is the most taxon-rich phylogenetic investigation of Tor and Neolissochilus, with 76% and 53% of species diversity, respectively. We evaluated the phylogenetic relationships of Tor and Neolissochilus, revealed the rich species diversity in Southeast Asia, and explored the species differentiation and distribution patterns. Broad trophic polymorphism, overlapping distribution zones, and minor interspecific genetic variations further complicate species identification and classification. Comprehensive sampling and precision analysis of morphological as well as omics data can better provide a perspective for further research on the origin, distribution and evolution of these taxa in Southeast Asia.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors’ Contributions
Ling Wu was responsible for investigation, methodology, data curation, software, formal analysis, visualization, and writing—original draft. Tao Qin was responsible for conceptualization, investigation, and writing—review and editing. Huy Duc Hoang was responsible for resources and writing—review and editing. Thaung Naing Oo was responsible for project administration and funding acquisition. Xiao-Ai Wang was responsible for validation and resources. Xiao-Yong Chen was responsible for conceptualization, methodology, project administration, supervision, validation, and funding acquisition, resources.
Acknowledgments
This work was supported by the Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01), the International Partnership Program of Chinese Academy of Sciences: Transboundary Cooperation on Biodiversity Research and Conservation in Gaoligong Mountains (E1ZK251), Yunnan Province Science and Technology Department (202203AP140007), and Lancang-Mekong Cooperation (LMC) Special Fund (Biodiversity Monitoring and Network Construction along Lancang-Mekong River Basin Project) to Xiao-Yong Chen; the Vietnam National Foundation for Science and Technology Development (NAFOSTEd-grant number 106.06-2017.40) to Huy Duc Hoang. We are grateful to Zhi-Bang Wang, Xin-Yuan Song, Feng Lin, and Khin Yadanar Htay for their help during this research.
Open Research
Data Availability
The new sequences of COI, Cyt b, 16S, and ND4 that support the findings of this study are deposited in the GenBank database.




